What is JupyterLab?¶
interactive python/R programming env
use the computationable power from our HPC
access the HPC data directly
Common usage¶
create a python/R notebook
create table of content and cell tags
[navigation] go to my data dir and start analysis
is my notebook running?
Data analysis using Pandas¶
read_csv(), head(), sample(), shape(), columns
df.isnull().any().any()
sort_values()
value_counts()
describe()
groupby()
min(),max(),std(),median()
subset dataframe
to_csv()
Example Data: several clinicl measurements of 48 Type 2 diabetes patients and 48 normal individuals¶
[22]:
import pandas as pd
df = pd.read_csv("https://raw.githubusercontent.com/YichaoOU/Data_Science_Tutorials/master/LTC_selected_features.csv",sep=",")
read data¶
[5]:
df.head()
[5]:
| SampleID | Peptide_27 | Fasting_plasma_glucose_(mmol/l) | HbA1c | Fasting_plasma_insulin_(pmol/l) | C-Peptide_(nmol/l) | HOMA-IR | Free_fatty_acids_(mmol/l) | Class | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 10.42 | 4.73 | 6.5 | 121.4 | 1.63 | 3.7 | 1.03 | 1 |
| 1 | 5 | 102.86 | 7.16 | 7.6 | 226.0 | 1.11 | 10.4 | 0.98 | 1 |
| 2 | 6 | 9.84 | 5.06 | 4.9 | 39.7 | 0.75 | 1.3 | 0.35 | 0 |
| 3 | 7 | 41.03 | 5.35 | 6.3 | 203.3 | 2.02 | 7.0 | 1.17 | 1 |
| 4 | 8 | 12.30 | 5.37 | 5.6 | 50.8 | 1.19 | 1.7 | 0.32 | 0 |
[6]:
df.sample(n=3)
[6]:
| SampleID | Peptide_27 | Fasting_plasma_glucose_(mmol/l) | HbA1c | Fasting_plasma_insulin_(pmol/l) | C-Peptide_(nmol/l) | HOMA-IR | Free_fatty_acids_(mmol/l) | Class | |
|---|---|---|---|---|---|---|---|---|---|
| 9 | 13 | 27.82 | 5.67 | 5.8 | 79.8 | 1.27 | 2.9 | 0.19 | 0 |
| 88 | 92 | 11.74 | 4.90 | 5.0 | 135.3 | 1.04 | 4.2 | 0.28 | 0 |
| 60 | 64 | 114.61 | 18.74 | 10.5 | 207.0 | 1.00 | 24.8 | 1.16 | 1 |
[25]:
df.columns
[25]:
Index(['SampleID', 'Peptide_27', 'Fasting_plasma_glucose_(mmol/l)', 'HbA1c',
'Fasting_plasma_insulin_(pmol/l)', 'C-Peptide_(nmol/l)', 'HOMA-IR',
'Free_fatty_acids_(mmol/l)', 'Class'],
dtype='object')
[7]:
df.isnull().any().any()
[7]:
False
get some statistics from data¶
[8]:
df.describe()
[8]:
| SampleID | Peptide_27 | Fasting_plasma_glucose_(mmol/l) | HbA1c | Fasting_plasma_insulin_(pmol/l) | C-Peptide_(nmol/l) | HOMA-IR | Free_fatty_acids_(mmol/l) | Class | |
|---|---|---|---|---|---|---|---|---|---|
| count | 96.000000 | 96.000000 | 96.000000 | 96.000000 | 96.000000 | 96.000000 | 96.000000 | 96.000000 | 96.000000 |
| mean | 51.541667 | 33.570208 | 6.711146 | 6.248958 | 155.566667 | 1.488632 | 7.240625 | 0.630937 | 0.500000 |
| std | 27.961596 | 23.398125 | 2.734147 | 1.455515 | 141.817693 | 0.778423 | 7.617384 | 0.343940 | 0.502625 |
| min | 3.000000 | 2.630000 | 3.800000 | 4.300000 | 15.000000 | 0.310000 | 0.500000 | 0.150000 | 0.000000 |
| 25% | 27.750000 | 15.597500 | 5.037500 | 5.300000 | 64.550000 | 0.935000 | 2.200000 | 0.340000 | 0.000000 |
| 50% | 51.500000 | 27.280000 | 5.670000 | 5.750000 | 112.600000 | 1.235000 | 4.100000 | 0.550000 | 0.500000 |
| 75% | 75.250000 | 46.885000 | 7.352500 | 6.625000 | 199.775000 | 1.845000 | 10.125000 | 0.940000 | 1.000000 |
| max | 100.000000 | 114.610000 | 18.740000 | 10.900000 | 783.000000 | 4.180000 | 45.300000 | 1.350000 | 1.000000 |
[9]:
df = df.sort_values("HbA1c",ascending=False)
[10]:
df.head()
[10]:
| SampleID | Peptide_27 | Fasting_plasma_glucose_(mmol/l) | HbA1c | Fasting_plasma_insulin_(pmol/l) | C-Peptide_(nmol/l) | HOMA-IR | Free_fatty_acids_(mmol/l) | Class | |
|---|---|---|---|---|---|---|---|---|---|
| 55 | 59 | 55.81 | 9.71 | 10.9 | 212.0 | 2.26 | 13.2 | 0.39 | 1 |
| 19 | 23 | 53.11 | 8.24 | 10.8 | 242.0 | 2.42 | 12.8 | 0.83 | 1 |
| 62 | 66 | 23.47 | 10.13 | 10.7 | 143.1 | 1.02 | 9.3 | 0.92 | 1 |
| 60 | 64 | 114.61 | 18.74 | 10.5 | 207.0 | 1.00 | 24.8 | 1.16 | 1 |
| 85 | 89 | 56.46 | 9.89 | 9.1 | 178.2 | 0.94 | 11.3 | 1.17 | 1 |
[12]:
df.value_counts('Class')
[12]:
Class
0 48
1 48
dtype: int64
[13]:
df.shape
[13]:
(96, 9)
[14]:
df['HbA1c'].min()
[14]:
4.3
[15]:
df['HbA1c'].median()
[15]:
5.75
[42]:
df['HbA1c'].max()
[42]:
10.9
[16]:
df.groupby('Class').mean()
[16]:
| SampleID | Peptide_27 | Fasting_plasma_glucose_(mmol/l) | HbA1c | Fasting_plasma_insulin_(pmol/l) | C-Peptide_(nmol/l) | HOMA-IR | Free_fatty_acids_(mmol/l) | |
|---|---|---|---|---|---|---|---|---|
| Class | ||||||||
| 0 | 53.354167 | 21.965417 | 5.321667 | 5.345833 | 79.208333 | 1.103513 | 2.679167 | 0.391042 |
| 1 | 49.729167 | 45.175000 | 8.100625 | 7.152083 | 231.925000 | 1.873750 | 11.802083 | 0.870833 |
[21]:
df.groupby('Class').head()
[21]:
| SampleID | Peptide_27 | Fasting_plasma_glucose_(mmol/l) | HbA1c | Fasting_plasma_insulin_(pmol/l) | C-Peptide_(nmol/l) | HOMA-IR | Free_fatty_acids_(mmol/l) | Class | |
|---|---|---|---|---|---|---|---|---|---|
| 55 | 59 | 55.81 | 9.71 | 10.9 | 212.0 | 2.26 | 13.2 | 0.39 | 1 |
| 19 | 23 | 53.11 | 8.24 | 10.8 | 242.0 | 2.42 | 12.8 | 0.83 | 1 |
| 62 | 66 | 23.47 | 10.13 | 10.7 | 143.1 | 1.02 | 9.3 | 0.92 | 1 |
| 60 | 64 | 114.61 | 18.74 | 10.5 | 207.0 | 1.00 | 24.8 | 1.16 | 1 |
| 85 | 89 | 56.46 | 9.89 | 9.1 | 178.2 | 0.94 | 11.3 | 1.17 | 1 |
| 89 | 93 | 18.06 | 7.39 | 6.5 | 88.9 | 1.22 | 4.2 | 0.35 | 0 |
| 65 | 69 | 43.86 | 6.26 | 6.0 | 69.2 | 1.20 | 2.8 | 0.22 | 0 |
| 43 | 47 | 11.36 | 6.67 | 5.9 | 58.2 | 0.89 | 2.5 | 0.36 | 0 |
| 35 | 39 | 32.81 | 5.68 | 5.9 | 102.4 | 1.03 | 3.7 | 0.27 | 0 |
| 6 | 10 | 25.75 | 5.67 | 5.8 | 67.1 | 0.94 | 2.4 | 0.26 | 0 |
[41]:
df.groupby('Class').head().to_csv("My_examlpe.tsv",index=False,sep="\t")
subset data¶
[60]:
df_undiagnosed = df[df['HbA1c']<6.5]
[61]:
df_undiagnosed['Class'].value_counts()
[61]:
0 47
1 18
Name: Class, dtype: int64
[62]:
df_undiagnosed['Class'].value_counts(normalize=True)
[62]:
0 0.723077
1 0.276923
Name: Class, dtype: float64
[26]:
data2 = "https://raw.githubusercontent.com/YichaoOU/Data_Science_Tutorials/master/newly_diagnosed.csv"
Exercise¶
Read this table as Pandas object¶
[36]:
df2 = pd.read_csv(data2,sep=",")
[38]:
df2.head()
[38]:
| Sample ID | Peptide 1 | Peptide 2 | Peptide 3 | Peptide 4 | Peptide 5 | Peptide 6 | Peptide 7 | Peptide 8 | Peptide 9 | ... | TSH (mU/l) | fT3 (pmol/l) | fT4 (pmol/l) | Cortisol (nmol/l) | Testosteron (nmol/l) | HOMA-IR | Free fatty acids (mmol/l) | RRsys (mmHg) | RR dia (mmHg) | ssCRP (mg/dl) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | sample 2 | 33.58 | 7.18 | 9.35 | 3.57 | 94.44 | 14.91 | 153.05 | 35.52 | 9.76 | ... | 0.72 | 5.23 | 21.5 | NaN | NaN | 7.2 | 0.30 | NaN | NaN | 0.48 |
| 1 | sample 3 | 37.57 | 8.70 | 10.79 | 3.36 | 94.11 | 15.99 | 198.88 | 39.65 | 8.62 | ... | 0.97 | NaN | NaN | NaN | NaN | 9.3 | 1.04 | NaN | NaN | 7.15 |
| 2 | sample 4 | 27.31 | 5.42 | 5.64 | 2.75 | 67.01 | 11.91 | 148.32 | 28.90 | 5.64 | ... | 1.02 | 5.01 | 17.6 | NaN | NaN | 9.1 | 0.37 | NaN | NaN | NaN |
| 3 | sample 5 | 29.09 | 5.81 | 4.69 | 3.61 | 65.99 | 12.42 | 154.55 | 26.48 | 6.38 | ... | 1.00 | 4.61 | 15.7 | NaN | NaN | 24.5 | 0.99 | 168.0 | 95.0 | 0.20 |
| 4 | sample 6 | 41.13 | 8.40 | 8.85 | 3.56 | 109.13 | 17.60 | 209.62 | 44.35 | 10.43 | ... | 1.30 | 5.43 | 19.2 | NaN | NaN | 11.1 | 0.54 | 165.0 | 84.0 | 0.30 |
5 rows × 69 columns
What are the features (columns) in this dataset?¶
[40]:
df2.columns
[40]:
Index(['Sample ID', 'Peptide 1', 'Peptide 2', 'Peptide 3', 'Peptide 4',
'Peptide 5', 'Peptide 6', 'Peptide 7', 'Peptide 8', 'Peptide 9',
'Peptide 10', 'Peptide 11', 'Peptide 12', 'Peptide 13', 'Peptide 14',
'Peptide 15', 'Peptide 16', 'Peptide 17', 'Peptide 18', 'Peptide 21',
'Peptide 22', 'Peptide 23', 'Peptide 24', 'Peptide 25', 'Peptide 26',
'Peptide 27', 'Peptide 29', 'Peptide 30', 'Age', 'Diagnosis', 'BMI',
'HbA1c (%)', 'Gender', 'Height', 'Body weight', 'BMI.1', 'Body fat',
'Fat free mass', 'Waist', 'Hip', 'WHR', 'Hemoglobin', 'Erythrozyten',
'Thrombozyten', 'Leukocytes', 'ALAT', 'ASAT', 'gGT',
'Fasting plasma glucose (mmol/l)', 'Fasting plasma insulin (pmol/l)',
'C-Peptide (nmol/l)', 'Proinsulin (pmol/l)', 'Creatinin',
'Triglycerides (mmol/l)', 'Cholesterol total (mmol/l)',
'HDL-Cholesterol (mmol/l)', 'LDL-Cholesterol (mmol/l)',
'Proteins total (g/l)', 'Albumin (g/l)', 'TSH (mU/l)', 'fT3 (pmol/l)',
'fT4 (pmol/l)', 'Cortisol (nmol/l)', 'Testosteron (nmol/l)', 'HOMA-IR',
'Free fatty acids (mmol/l)', 'RRsys (mmHg)', 'RR dia (mmHg)',
'ssCRP (mg/dl)'],
dtype='object')
What are the maximum and minimum values for HbA1c? Are they the same as the first dataset?¶
[45]:
df2['HbA1c (%)'].min()
[45]:
4.3
[46]:
df2['HbA1c (%)'].max()
[46]:
9.7
Does our data have NaN values?¶
[47]:
df2.isnull().any().any()
[47]:
True
[52]:
df2[df2.isnull().any(axis=1)] # any rows containing NaN
[52]:
| Sample ID | Peptide 1 | Peptide 2 | Peptide 3 | Peptide 4 | Peptide 5 | Peptide 6 | Peptide 7 | Peptide 8 | Peptide 9 | ... | TSH (mU/l) | fT3 (pmol/l) | fT4 (pmol/l) | Cortisol (nmol/l) | Testosteron (nmol/l) | HOMA-IR | Free fatty acids (mmol/l) | RRsys (mmHg) | RR dia (mmHg) | ssCRP (mg/dl) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | sample 2 | 33.58 | 7.18 | 9.35 | 3.57 | 94.44 | 14.91 | 153.05 | 35.52 | 9.76 | ... | 0.72 | 5.23 | 21.5 | NaN | NaN | 7.2 | 0.30 | NaN | NaN | 0.48 |
| 1 | sample 3 | 37.57 | 8.70 | 10.79 | 3.36 | 94.11 | 15.99 | 198.88 | 39.65 | 8.62 | ... | 0.97 | NaN | NaN | NaN | NaN | 9.3 | 1.04 | NaN | NaN | 7.15 |
| 2 | sample 4 | 27.31 | 5.42 | 5.64 | 2.75 | 67.01 | 11.91 | 148.32 | 28.90 | 5.64 | ... | 1.02 | 5.01 | 17.6 | NaN | NaN | 9.1 | 0.37 | NaN | NaN | NaN |
| 3 | sample 5 | 29.09 | 5.81 | 4.69 | 3.61 | 65.99 | 12.42 | 154.55 | 26.48 | 6.38 | ... | 1.00 | 4.61 | 15.7 | NaN | NaN | 24.5 | 0.99 | 168.0 | 95.0 | 0.20 |
| 4 | sample 6 | 41.13 | 8.40 | 8.85 | 3.56 | 109.13 | 17.60 | 209.62 | 44.35 | 10.43 | ... | 1.30 | 5.43 | 19.2 | NaN | NaN | 11.1 | 0.54 | 165.0 | 84.0 | 0.30 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 91 | Sample 96 | 26.08 | 3.54 | 0.68 | 0.05 | 29.46 | 9.27 | 142.70 | 18.61 | 2.49 | ... | 3.84 | NaN | NaN | 20.30 | 117.00 | 0.8 | 0.44 | 130.0 | 80.0 | 5.50 |
| 92 | Sample 97 | 17.72 | 2.28 | 0.64 | 0.01 | 19.22 | 7.37 | 102.24 | 13.22 | 1.61 | ... | 0.63 | NaN | NaN | 9.73 | 208.00 | 5.0 | 0.36 | NaN | NaN | NaN |
| 93 | Sample 98 | 17.70 | 2.23 | 0.59 | 0.01 | 18.47 | 5.80 | 103.94 | 13.67 | 1.65 | ... | 1.36 | NaN | NaN | 9.03 | 56.00 | 2.7 | 0.39 | NaN | NaN | NaN |
| 94 | Sample 99 | 26.57 | 3.33 | 0.71 | 0.07 | 27.69 | 8.31 | 137.81 | 20.68 | 2.99 | ... | 0.71 | NaN | NaN | NaN | 1.49 | 4.6 | 0.76 | NaN | NaN | 0.08 |
| 95 | Sample 100 | 25.26 | 3.40 | 0.64 | 0.05 | 27.47 | 8.72 | 138.50 | 19.72 | 2.54 | ... | 3.34 | NaN | NaN | 11.50 | 93.00 | 4.7 | 0.35 | NaN | NaN | 19.50 |
96 rows × 69 columns
[56]:
df2[df2.columns[df2.isnull().any(axis=0)]] # any columns containing NaN
[56]:
| Body fat | Fat free mass | Waist | Hip | WHR | Hemoglobin | Erythrozyten | Thrombozyten | Leukocytes | ALAT | ... | Proteins total (g/l) | Albumin (g/l) | TSH (mU/l) | fT3 (pmol/l) | fT4 (pmol/l) | Cortisol (nmol/l) | Testosteron (nmol/l) | RRsys (mmHg) | RR dia (mmHg) | ssCRP (mg/dl) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 32.7 | 73.4 | NaN | NaN | NaN | 8.3 | 4.32 | 216.0 | 7.4 | 0.29 | ... | 70.8 | NaN | 0.72 | 5.23 | 21.5 | NaN | NaN | NaN | NaN | 0.48 |
| 1 | 33.9 | NaN | NaN | NaN | NaN | 8.9 | 5.44 | 180.0 | 5.9 | 1.40 | ... | NaN | NaN | 0.97 | NaN | NaN | NaN | NaN | NaN | NaN | 7.15 |
| 2 | 33.8 | NaN | NaN | NaN | NaN | 8.9 | 4.68 | 169.0 | 5.4 | 0.55 | ... | 66.1 | NaN | 1.02 | 5.01 | 17.6 | NaN | NaN | NaN | NaN | NaN |
| 3 | NaN | NaN | NaN | NaN | NaN | 8.5 | 4.83 | 237.0 | 6.6 | 0.57 | ... | 72.1 | NaN | 1.00 | 4.61 | 15.7 | NaN | NaN | 168.0 | 95.0 | 0.20 |
| 4 | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | 0.65 | ... | 76.3 | NaN | 1.30 | 5.43 | 19.2 | NaN | NaN | 165.0 | 84.0 | 0.30 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 91 | 28.0 | 73.6 | NaN | NaN | NaN | 10.1 | 5.28 | 170.0 | 8.5 | 0.65 | ... | 67.9 | 43.6 | 3.84 | NaN | NaN | 20.30 | 117.00 | 130.0 | 80.0 | 5.50 |
| 92 | 37.9 | 81.2 | NaN | NaN | NaN | 7.4 | 4.18 | 395.0 | 10.1 | 0.30 | ... | NaN | NaN | 0.63 | NaN | NaN | 9.73 | 208.00 | NaN | NaN | NaN |
| 93 | 36.8 | 79.5 | NaN | NaN | NaN | 9.1 | 4.76 | 172.0 | 7.8 | 0.43 | ... | NaN | NaN | 1.36 | NaN | NaN | 9.03 | 56.00 | NaN | NaN | NaN |
| 94 | 46.9 | 66.7 | 135.0 | 137.0 | 0.99 | 8.3 | 4.84 | 196.0 | 5.9 | 0.43 | ... | NaN | NaN | 0.71 | NaN | NaN | NaN | 1.49 | NaN | NaN | 0.08 |
| 95 | 30.6 | 90.4 | NaN | NaN | NaN | 8.5 | 4.61 | 219.0 | 3.7 | 1.16 | ... | NaN | NaN | 3.34 | NaN | NaN | 11.50 | 93.00 | NaN | NaN | 19.50 |
96 rows × 28 columns
How many diabetic patents will be undiagnosed using HbA1c < 6.5?¶
[64]:
df2_undiagnosed = df2[df2['HbA1c (%)']<6.5]
[67]:
df2_undiagnosed['Diagnosis'].value_counts()
[67]:
NGT 48
T2D 23
Name: Diagnosis, dtype: int64
[68]:
df2_undiagnosed['Diagnosis'].value_counts(normalize=True)
[68]:
NGT 0.676056
T2D 0.323944
Name: Diagnosis, dtype: float64
Save the undiagnosed table as “happy_learning.csv”¶
Data visualization using Seaborn and many other libraries¶
scatter plot
barplot
boxplot
violin plot
beeswarm plot
Scatter plots¶
[69]:
import seaborn as sns
[71]:
df.columns
[71]:
Index(['SampleID', 'Peptide_27', 'Fasting_plasma_glucose_(mmol/l)', 'HbA1c',
'Fasting_plasma_insulin_(pmol/l)', 'C-Peptide_(nmol/l)', 'HOMA-IR',
'Free_fatty_acids_(mmol/l)', 'Class'],
dtype='object')
[72]:
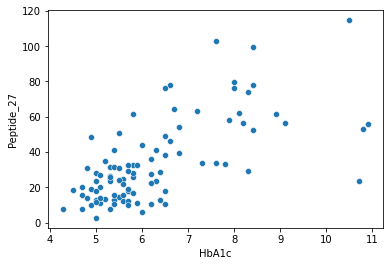
sns.scatterplot(data = df,x="HbA1c",y="Peptide_27")
[72]:
<AxesSubplot:xlabel='HbA1c', ylabel='Peptide_27'>
[73]:
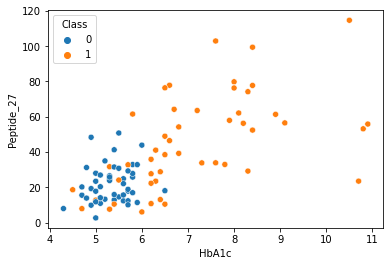
sns.scatterplot(data = df,x="HbA1c",y="Peptide_27",hue="Class")
[73]:
<AxesSubplot:xlabel='HbA1c', ylabel='Peptide_27'>
[74]:
import matplotlib.pylab as plt
[76]:
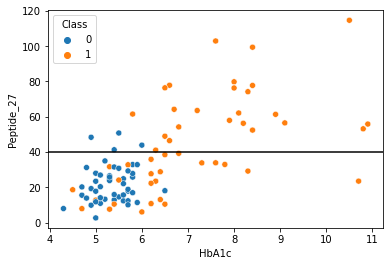
sns.scatterplot(data = df,x="HbA1c",y="Peptide_27",hue="Class")
plt.axhline(40,color="black")
[76]:
<matplotlib.lines.Line2D at 0x2aad6a85d6d0>
[78]:
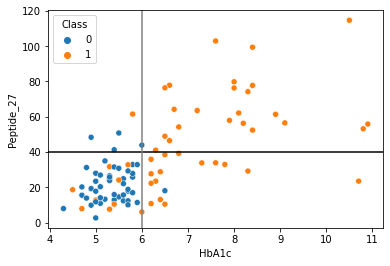
sns.scatterplot(data = df,x="HbA1c",y="Peptide_27",hue="Class")
plt.axhline(40,color="black")
plt.axvline(6,color="grey")
[78]:
<matplotlib.lines.Line2D at 0x2aad6aa23130>
[124]:
import plotly.express as px
import plotly.io as pio
pio.renderers.default = "iframe"
[140]:
df.head()
[140]:
| SampleID | Peptide_27 | Fasting_plasma_glucose_(mmol/l) | HbA1c | Fasting_plasma_insulin_(pmol/l) | C-Peptide_(nmol/l) | HOMA-IR | Free_fatty_acids_(mmol/l) | Class | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 10.42 | 4.73 | 6.5 | 121.4 | 1.63 | 3.7 | 1.03 | 1 |
| 1 | 5 | 102.86 | 7.16 | 7.6 | 226.0 | 1.11 | 10.4 | 0.98 | 1 |
| 2 | 6 | 9.84 | 5.06 | 4.9 | 39.7 | 0.75 | 1.3 | 0.35 | 0 |
| 3 | 7 | 41.03 | 5.35 | 6.3 | 203.3 | 2.02 | 7.0 | 1.17 | 1 |
| 4 | 8 | 12.30 | 5.37 | 5.6 | 50.8 | 1.19 | 1.7 | 0.32 | 0 |
interactive scatter plot¶
[142]:
px.scatter(data_frame = df,x="HbA1c",y="Peptide_27",color="Class",hover_data=["SampleID"])
[79]:
df.describe()
[79]:
| SampleID | Peptide_27 | Fasting_plasma_glucose_(mmol/l) | HbA1c | Fasting_plasma_insulin_(pmol/l) | C-Peptide_(nmol/l) | HOMA-IR | Free_fatty_acids_(mmol/l) | Class | |
|---|---|---|---|---|---|---|---|---|---|
| count | 96.000000 | 96.000000 | 96.000000 | 96.000000 | 96.000000 | 96.000000 | 96.000000 | 96.000000 | 96.000000 |
| mean | 51.541667 | 33.570208 | 6.711146 | 6.248958 | 155.566667 | 1.488632 | 7.240625 | 0.630937 | 0.500000 |
| std | 27.961596 | 23.398125 | 2.734147 | 1.455515 | 141.817693 | 0.778423 | 7.617384 | 0.343940 | 0.502625 |
| min | 3.000000 | 2.630000 | 3.800000 | 4.300000 | 15.000000 | 0.310000 | 0.500000 | 0.150000 | 0.000000 |
| 25% | 27.750000 | 15.597500 | 5.037500 | 5.300000 | 64.550000 | 0.935000 | 2.200000 | 0.340000 | 0.000000 |
| 50% | 51.500000 | 27.280000 | 5.670000 | 5.750000 | 112.600000 | 1.235000 | 4.100000 | 0.550000 | 0.500000 |
| 75% | 75.250000 | 46.885000 | 7.352500 | 6.625000 | 199.775000 | 1.845000 | 10.125000 | 0.940000 | 1.000000 |
| max | 100.000000 | 114.610000 | 18.740000 | 10.900000 | 783.000000 | 4.180000 | 45.300000 | 1.350000 | 1.000000 |
barplot¶
[83]:
plot_df = df.melt(id_vars=['Class'],value_vars = ['Fasting_plasma_glucose_(mmol/l)','HbA1c'])
sns.barplot(data=plot_df,x="variable",y="value",hue="Class")
[83]:
<AxesSubplot:xlabel='variable', ylabel='value'>
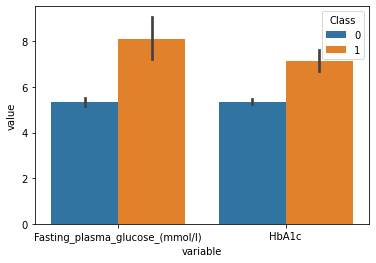
[166]:
df_norm = df/df.max(axis=0)
df_norm.head()
[166]:
| SampleID | Peptide_27 | Fasting_plasma_glucose_(mmol/l) | HbA1c | Fasting_plasma_insulin_(pmol/l) | C-Peptide_(nmol/l) | HOMA-IR | Free_fatty_acids_(mmol/l) | Class | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.03 | 0.090917 | 0.252401 | 0.596330 | 0.155045 | 0.389952 | 0.081678 | 0.762963 | 1.0 |
| 1 | 0.05 | 0.897478 | 0.382070 | 0.697248 | 0.288633 | 0.265550 | 0.229581 | 0.725926 | 1.0 |
| 2 | 0.06 | 0.085856 | 0.270011 | 0.449541 | 0.050702 | 0.179426 | 0.028698 | 0.259259 | 0.0 |
| 3 | 0.07 | 0.357997 | 0.285486 | 0.577982 | 0.259642 | 0.483254 | 0.154525 | 0.866667 | 1.0 |
| 4 | 0.08 | 0.107320 | 0.286553 | 0.513761 | 0.064879 | 0.284689 | 0.037528 | 0.237037 | 0.0 |
[175]:
plot_df = df_norm.melt(id_vars=['Class'],value_vars =list(set(df_norm.columns)-set(['Class','SampleID'])) )
sns.barplot(data=plot_df,y="variable",x="value",hue="Class")
[175]:
<AxesSubplot:xlabel='value', ylabel='variable'>
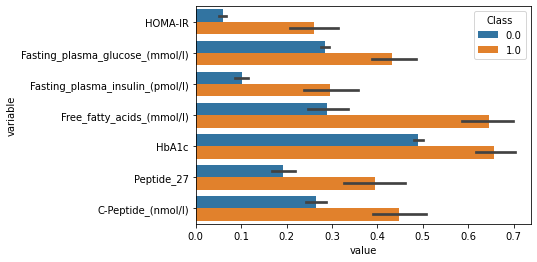
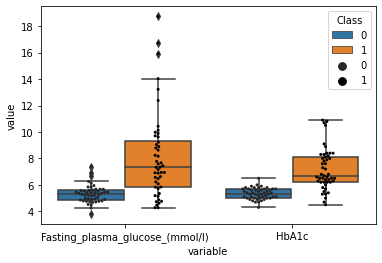
boxplot¶
[84]:
sns.boxplot(data=plot_df,x="variable",y="value",hue="Class")
[84]:
<AxesSubplot:xlabel='variable', ylabel='value'>
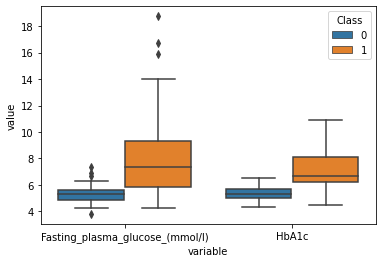
Violinplot¶
[86]:
sns.violinplot(data=plot_df,x="variable",y="value",hue="Class")
[86]:
<AxesSubplot:xlabel='variable', ylabel='value'>
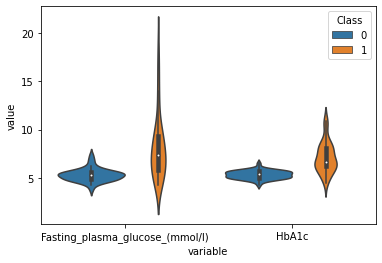
beeswarm plot¶
[89]:
sns.swarmplot(data=plot_df,x="variable",y="value",hue="Class")
[89]:
<AxesSubplot:xlabel='variable', ylabel='value'>
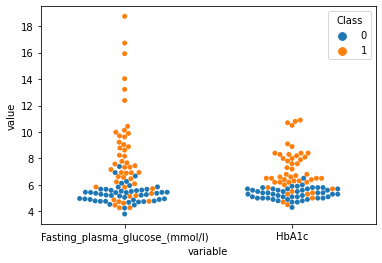
[100]:
sns.violinplot(data=plot_df,x="variable",y="value",hue="Class",inner=None)
sns.swarmplot(data=plot_df,x="variable",y="value",hue="Class",dodge=True,color="black",s=3)
[100]:
<AxesSubplot:xlabel='variable', ylabel='value'>

[101]:
sns.boxplot(data=plot_df,x="variable",y="value",hue="Class")
sns.swarmplot(data=plot_df,x="variable",y="value",hue="Class",dodge=True,color="black",s=3)
[101]:
<AxesSubplot:xlabel='variable', ylabel='value'>
[102]:
df.head()
[102]:
| SampleID | Peptide_27 | Fasting_plasma_glucose_(mmol/l) | HbA1c | Fasting_plasma_insulin_(pmol/l) | C-Peptide_(nmol/l) | HOMA-IR | Free_fatty_acids_(mmol/l) | Class | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 10.42 | 4.73 | 6.5 | 121.4 | 1.63 | 3.7 | 1.03 | 1 |
| 1 | 5 | 102.86 | 7.16 | 7.6 | 226.0 | 1.11 | 10.4 | 0.98 | 1 |
| 2 | 6 | 9.84 | 5.06 | 4.9 | 39.7 | 0.75 | 1.3 | 0.35 | 0 |
| 3 | 7 | 41.03 | 5.35 | 6.3 | 203.3 | 2.02 | 7.0 | 1.17 | 1 |
| 4 | 8 | 12.30 | 5.37 | 5.6 | 50.8 | 1.19 | 1.7 | 0.32 | 0 |
Exercise¶
Can we still use the same HbA1c and Peptide27 cutoff for the second data?¶
[160]:
df2.columns
[160]:
Index(['Sample ID', 'Peptide 1', 'Peptide 2', 'Peptide 3', 'Peptide 4',
'Peptide 5', 'Peptide 6', 'Peptide 7', 'Peptide 8', 'Peptide 9',
'Peptide 10', 'Peptide 11', 'Peptide 12', 'Peptide 13', 'Peptide 14',
'Peptide 15', 'Peptide 16', 'Peptide 17', 'Peptide 18', 'Peptide 21',
'Peptide 22', 'Peptide 23', 'Peptide 24', 'Peptide 25', 'Peptide 26',
'Peptide 27', 'Peptide 29', 'Peptide 30', 'Age', 'Diagnosis', 'BMI',
'HbA1c (%)', 'Gender', 'Height', 'Body weight', 'BMI.1', 'Body fat',
'Fat free mass', 'Waist', 'Hip', 'WHR', 'Hemoglobin', 'Erythrozyten',
'Thrombozyten', 'Leukocytes', 'ALAT', 'ASAT', 'gGT',
'Fasting plasma glucose (mmol/l)', 'Fasting plasma insulin (pmol/l)',
'C-Peptide (nmol/l)', 'Proinsulin (pmol/l)', 'Creatinin',
'Triglycerides (mmol/l)', 'Cholesterol total (mmol/l)',
'HDL-Cholesterol (mmol/l)', 'LDL-Cholesterol (mmol/l)',
'Proteins total (g/l)', 'Albumin (g/l)', 'TSH (mU/l)', 'fT3 (pmol/l)',
'fT4 (pmol/l)', 'Cortisol (nmol/l)', 'Testosteron (nmol/l)', 'HOMA-IR',
'Free fatty acids (mmol/l)', 'RRsys (mmHg)', 'RR dia (mmHg)',
'ssCRP (mg/dl)'],
dtype='object')
[161]:
sns.scatterplot(data = df2,x="HbA1c (%)",y="Peptide 27",hue="Diagnosis")
plt.axhline(40,color="black")
plt.axvline(6,color="grey")
[161]:
<matplotlib.lines.Line2D at 0x2aab879ffd90>
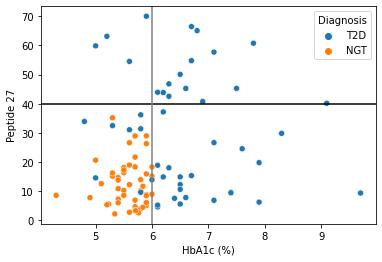
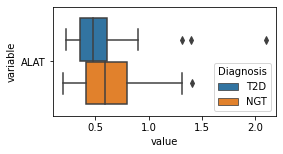
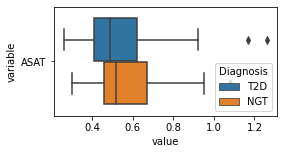
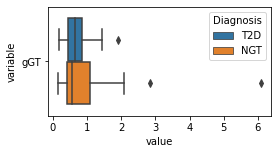
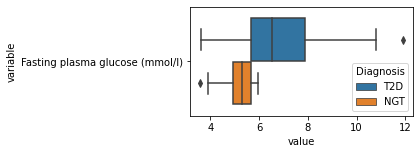
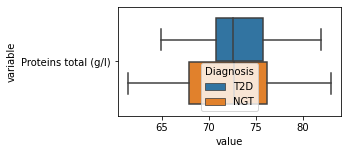
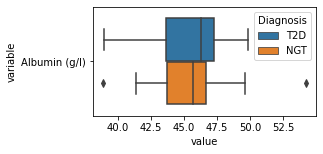
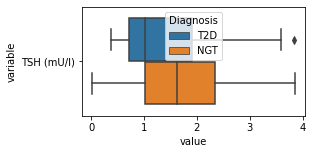
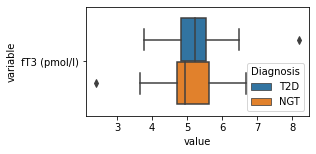
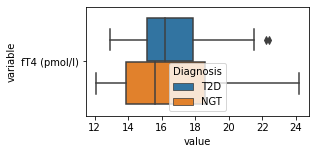
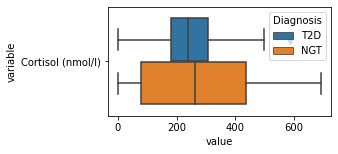
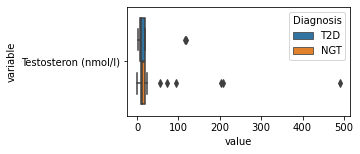
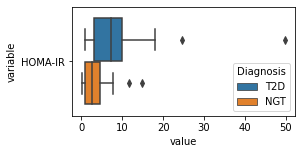
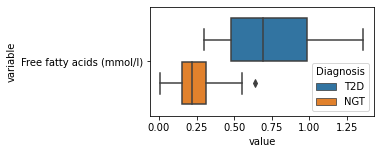
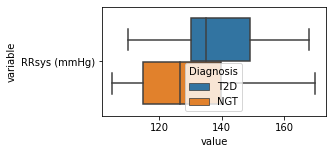
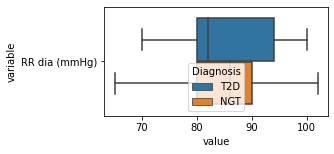
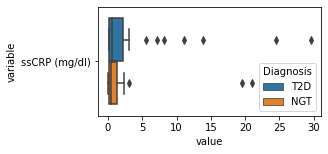
what is the data distribution for each feature?¶
[179]:
df2.head()
[179]:
| Sample ID | Peptide 1 | Peptide 2 | Peptide 3 | Peptide 4 | Peptide 5 | Peptide 6 | Peptide 7 | Peptide 8 | Peptide 9 | ... | TSH (mU/l) | fT3 (pmol/l) | fT4 (pmol/l) | Cortisol (nmol/l) | Testosteron (nmol/l) | HOMA-IR | Free fatty acids (mmol/l) | RRsys (mmHg) | RR dia (mmHg) | ssCRP (mg/dl) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | sample 2 | 33.58 | 7.18 | 9.35 | 3.57 | 94.44 | 14.91 | 153.05 | 35.52 | 9.76 | ... | 0.72 | 5.23 | 21.5 | NaN | NaN | 7.2 | 0.30 | NaN | NaN | 0.48 |
| 1 | sample 3 | 37.57 | 8.70 | 10.79 | 3.36 | 94.11 | 15.99 | 198.88 | 39.65 | 8.62 | ... | 0.97 | NaN | NaN | NaN | NaN | 9.3 | 1.04 | NaN | NaN | 7.15 |
| 2 | sample 4 | 27.31 | 5.42 | 5.64 | 2.75 | 67.01 | 11.91 | 148.32 | 28.90 | 5.64 | ... | 1.02 | 5.01 | 17.6 | NaN | NaN | 9.1 | 0.37 | NaN | NaN | NaN |
| 3 | sample 5 | 29.09 | 5.81 | 4.69 | 3.61 | 65.99 | 12.42 | 154.55 | 26.48 | 6.38 | ... | 1.00 | 4.61 | 15.7 | NaN | NaN | 24.5 | 0.99 | 168.0 | 95.0 | 0.20 |
| 4 | sample 6 | 41.13 | 8.40 | 8.85 | 3.56 | 109.13 | 17.60 | 209.62 | 44.35 | 10.43 | ... | 1.30 | 5.43 | 19.2 | NaN | NaN | 11.1 | 0.54 | 165.0 | 84.0 | 0.30 |
5 rows × 69 columns
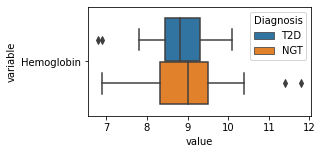
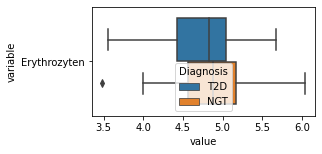
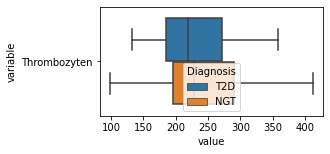
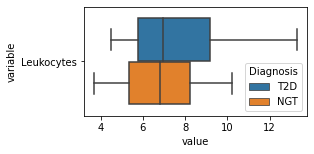
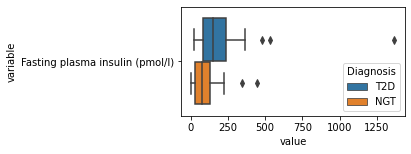
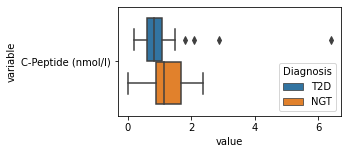
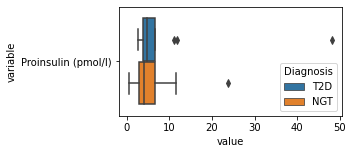
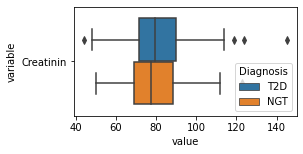
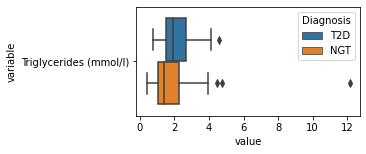
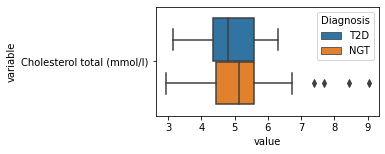
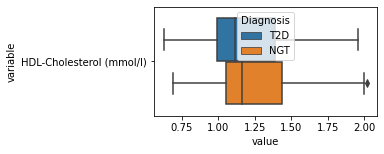
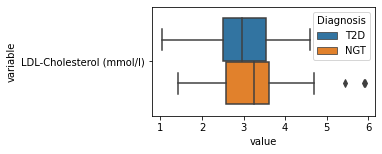
[200]:
for c in df2.columns:
try:
plt.figure(figsize=(4,2))
tmp = df2.melt(id_vars=['Diagnosis'],value_vars=c)
sns.boxplot(data=tmp,y="variable",x="value",hue="Diagnosis")
except:
continue
<ipython-input-200-795c994e6a81>:3: RuntimeWarning:
More than 20 figures have been opened. Figures created through the pyplot interface (`matplotlib.pyplot.figure`) are retained until explicitly closed and may consume too much memory. (To control this warning, see the rcParam `figure.max_open_warning`).
<Figure size 288x144 with 0 Axes>
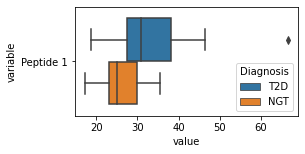
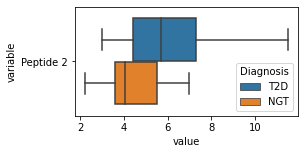
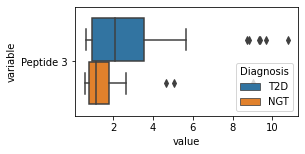
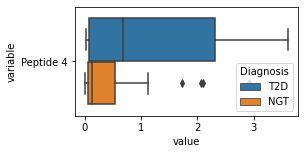
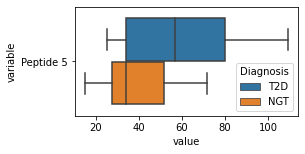
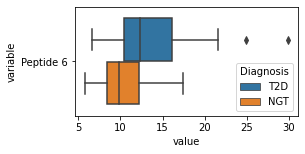
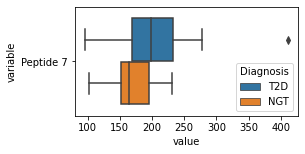
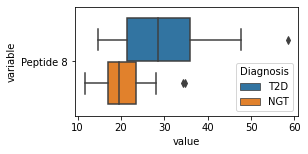
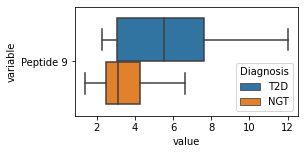
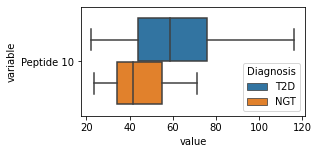
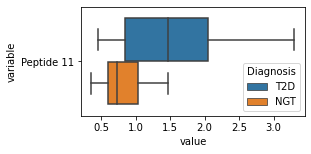
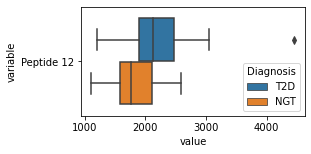
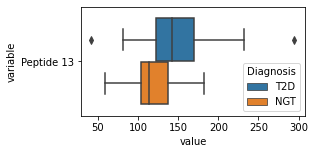
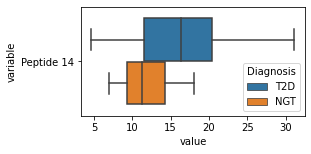
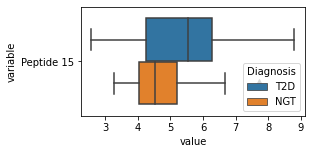
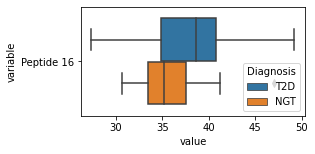
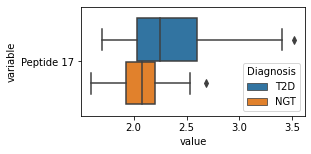
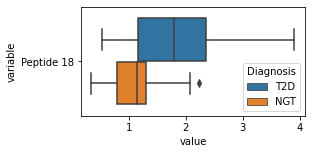
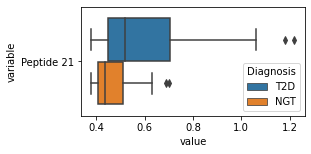
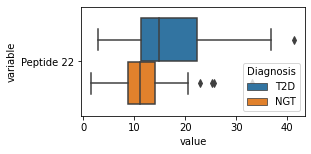
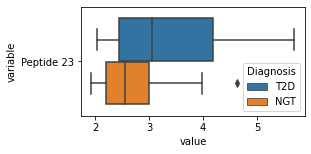
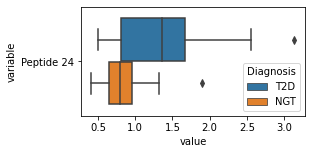
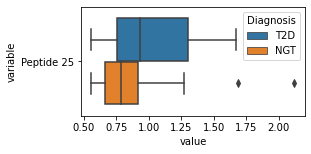
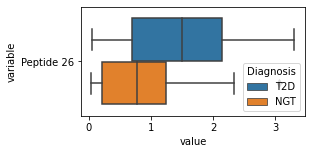
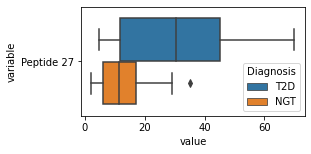
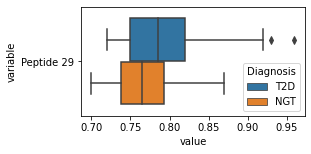
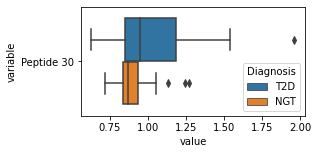
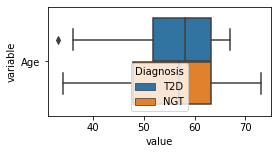
<Figure size 288x144 with 0 Axes>
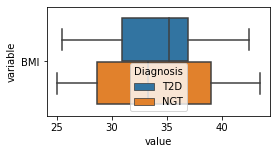
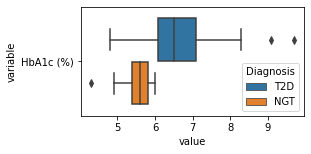
<Figure size 288x144 with 0 Axes>
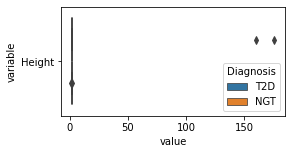
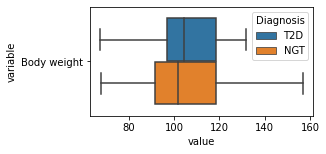
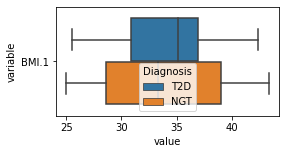
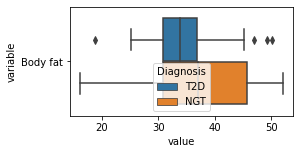
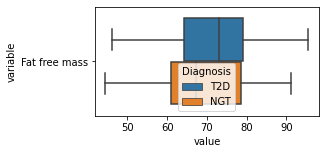
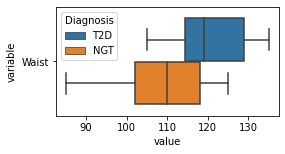
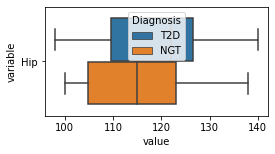
<Figure size 288x144 with 0 Axes>
[ ]: