Blood data enrichment test¶
Summary¶
This is a suite of tools for users to do an enrichment test given a list of bed files.
Input¶
A list of bed files. The first 3 columns of bed file should be chr, start, end.
Bed format¶
Additional columns are OK. The first 3 columns have to be chr, start, end.
chr11 4167364 4167385 chr11:4167374-4167375
chr11 4167366 4167387 chr11:4167376-4167377
chr11 4167367 4167388 chr11:4167377-4167378
chr11 4167370 4167391 chr11:4167380-4167381
Output¶
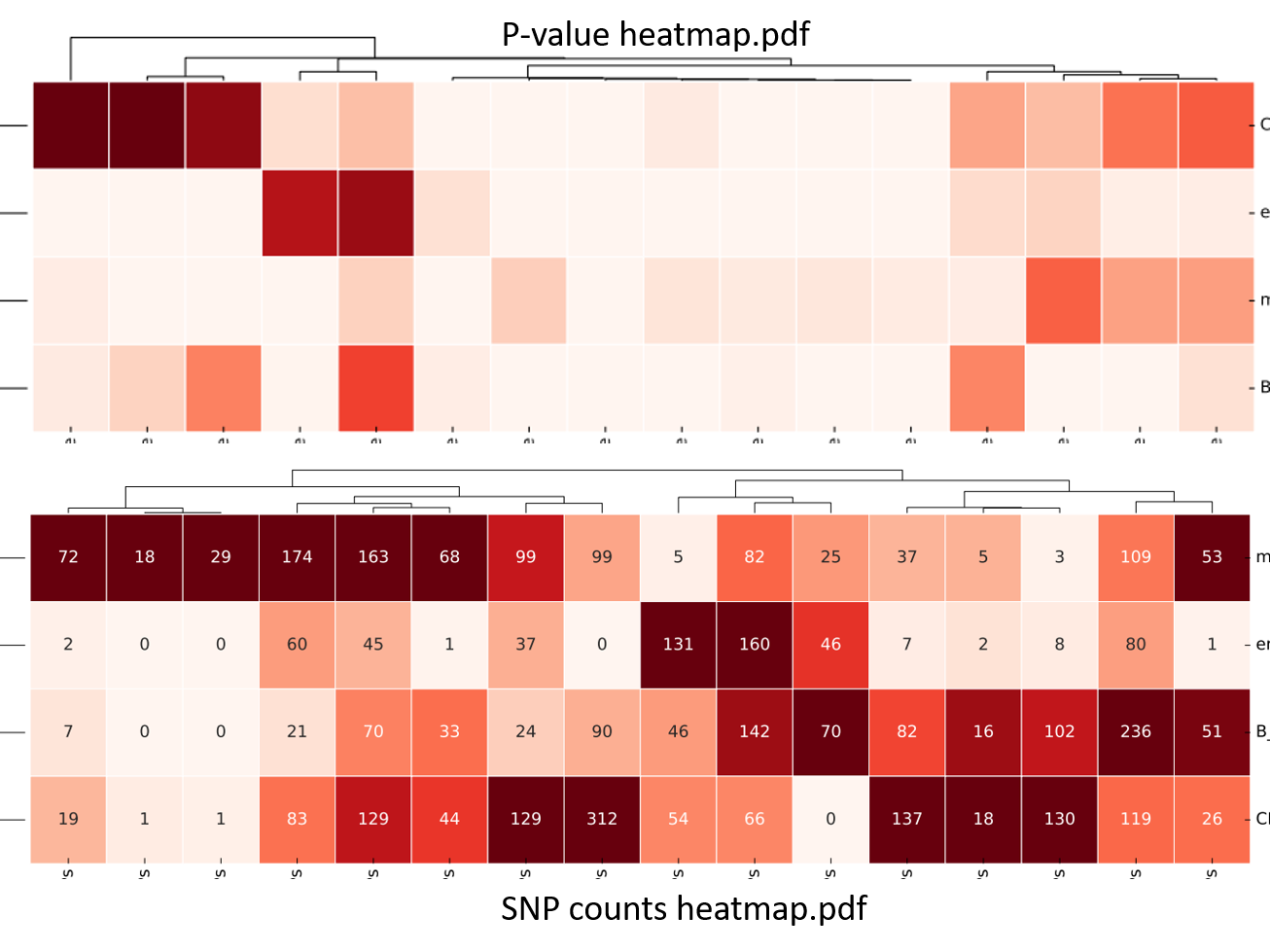
Blood traits variants/SNPs enrichment heatmap¶
Blood traits variants/SNPs enrichment ranked list¶
[yli11@nodecn262 summary_files]$ ls *.list
P_value_ranked.list SNP_count_ranked.list
Usage¶
Blood traits variants/SNPs enrichment¶
usage: GREGOR.py [-h] [-j JID] [-s SNP_LIST] [-n SNP_DATABASE_NAME] -f
BED_LIST [--template_config TEMPLATE_CONFIG]
optional arguments:
-h, --help show this help message and exit
-j JID, --jid JID enter a job ID, which is used to make a new directory.
Every output will be moved into this folder. (default:
GREGOR_yli11_2019-11-06)
-s SNP_LIST, --SNP_list SNP_LIST
Please provide absolute path if you use custom SNP
list (default: /research/rgs01/project_space/chenggrp/
blood_regulome/chenggrp/Data_resource/blood_traits/VJ_
2019/VJ01.list)
-n SNP_DATABASE_NAME, --SNP_database_name SNP_DATABASE_NAME
options are custom, hg19_gwas, VJ01_combined, VJ01,
VJ025, VJ05, VJ075 (default: VJ01)
-f BED_LIST, --bed_list BED_LIST
absolute or relative path (default: None)
--template_config TEMPLATE_CONFIG
hpcf_interactive
module load python/2.7.13
GREGOR.py -f bed.list -n VJ01
Note
You can provide your own SNP list using -n custom -s YOUR_list option. The SNP list file specifies the absolute paths to several trait-SNP lists. See below for an example of custom SNP list.
-s YOUR_list
You can have any number of list here.
-----YOUR_list------
/path/to/banana.list
actual SNP locations (e.g., banana.list)
-banana.list--
chr2:423434234
chr4:463421444