Feature extraction over a bed file¶
usage: annotate_peaks.py [-h] [-j JID] [--pipeline_type PIPELINE_TYPE]
[-w WINDOW_SIZE] [-s STEP_SIZE] [-n NUMBER_STEPS] -l
FEATURE_LIST [-t FEATURE_TYPE] [--bed_list BED_LIST]
[-g GENOME] -f INPUT
annotate your bed files given a list of bed files, bedGraph files, or
narrowPeak files. The input list can't contain a mix of types.
optional arguments:
-h, --help show this help message and exit
-j JID, --jid JID enter a job ID, which is used to make a new directory.
Every output will be moved into this folder. (default:
annotate_peaks_yli11_2019-07-13)
--pipeline_type PIPELINE_TYPE
Not for end-user. (default: annotate_peaks)
-w WINDOW_SIZE, --window_size WINDOW_SIZE
feature window size, use the center of you input bed
file, and create several windows both upstream and
downstream, then your input file list will be used to
overlap with each of these windows. (default: 200)
-s STEP_SIZE, --step_size STEP_SIZE
How to create each window (where to set window start
site). If step_size >= window_size, then it means no
overlap between each window. (default: 100)
-n NUMBER_STEPS, --number_steps NUMBER_STEPS
How many windows to create (i.e., how many steps you
want to go). Note that number of bins = n-1. The
actual bp to the center is n*s+w (default: 5)
-l FEATURE_LIST, --feature_list FEATURE_LIST
a tsv file containing 2 columns, feature name &
feature file (with path) (default: None)
-t FEATURE_TYPE, --feature_type FEATURE_TYPE
can be bed, bedGraph, or narrowPeak. Case insensitive.
Currently, only bedGraph or narrowPeak is implemented.
(default: narrowPeak)
--bed_list BED_LIST NOT FOR END-USER (default: None)
-g GENOME, --genome GENOME
homer genome, hg18, hg19, mm9, mm10. Case sensitive!
(default: hg19)
-f INPUT, --input INPUT
a bed file with at least 4 columns, additional columns
will be kept when output the result. The first 4
columns are chr, start, end, unique name.
(default: None)
Note
--feature_type bed option is not implemented.
Summary¶
Given a bed file, user wants to annotate this bed files with a list of bed files, bedGraph files, or narrowPeak files. In additional to regions in the user-input bed file, nearby region feature extraction will also be done by options -w, -s, and -n. Basically, those parameters are used to define a sliding window.
Note
the input feature list should not contain a mix of file types.
Flowchart¶
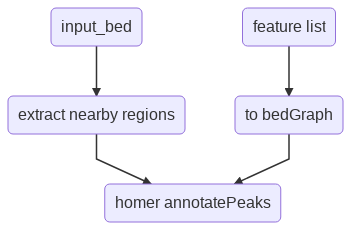
Input¶
A bed file to be annotated
This bed file must contain at least 4 columns: chr, start, end, unique_name. Additional columns will be kept when output the result.
Feature list
A tsv file, 2 columns: feature_name, feature_file (with path).
If feature_file is not in the current dir, you should provide relative or absolute path.
H2_ATAC /path_to_file/Hudep2_ATAC.narrowPeak
H1_ATAC /path_to_file/Hudep1_ATAC.narrowPeak
myGrandpa /path_to_file/myGrandpa_and_blue_color.narrowPeak
Usage¶
Go to your data directory and type the following.
Step 0: Load python version 2.7.13.
module load python/2.7.13
Step 1: Prepare input parameters
annotate_peaks.py -l feature.list -f gRNA.loci306.bed -g hg19
Note
By default, genome is hg19. Only hg18, hg19, mm9, mm10 is available for this option. Use -w, -s, and -n to control nearby windows.
Output¶
Once the job is finished, you will receive a notification email.
*_homer.tsv files contain the features overlaped with your input regions and nearby regions.
Comments¶
code @ github.