Find dense motif clusters¶
usage: motif_cluster.py [-h] [-j JID] -f INPUT_BED [-m MOTIF]
[--motif_cluster_score MOTIF_CLUSTER_SCORE]
[-o OUTPUT] [--input_list INPUT_LIST]
[--motif_matching_score MOTIF_MATCHING_SCORE]
[-g GENOME] [--genome_fasta GENOME_FASTA]
optional arguments:
-h, --help show this help message and exit
-j JID, --jid JID enter a job ID, which is used to make a new directory.
Every output will be moved into this folder. (default:
motif_cluster_yli11_2020-09-16)
-f INPUT_BED, --input_bed INPUT_BED
3 column bed file, additional columns are OK, but will
be ignored (default: None)
-m MOTIF, --motif MOTIF
motif database homer format (default: /home/yli11/Data
/Motif_database/homer/combined.homer.motif)
--motif_cluster_score MOTIF_CLUSTER_SCORE
motif_cluster_score (default: 5)
-o OUTPUT, --output OUTPUT
--input_list INPUT_LIST
not for end user (default: None)
--motif_matching_score MOTIF_MATCHING_SCORE
motif_matching_score (default: 6)
Genome Info:
-g GENOME, --genome GENOME
genome version: hg19, mm10, mm9, hg38 (default: hg19)
--genome_fasta GENOME_FASTA
genome version: hs, mm (default:
/home/yli11/Data/Human/hg19/fasta/hg19.fa)
Summary¶
This pipeline uses cluster bluster to find dense motif clusters.
Input¶
bed file
Usage¶
hpcf_interactive
module load python/2.7.13
motif_cluster.py -f input.bed -g mm9 -o output.bed
Output¶
The output is a bed file where the first 3 columns are the coordinates.
Motif cluster name or motif name is in the 4th column. Motif clusters are named as chr5:52866664-52868616__cluster_1, the individual motif in each cluster can be found in the lines below motif cluster.
5th column is motif cluster score or motif score.
6th column is the strand.
The genereated bedjs file can also be visualized in protein paint.
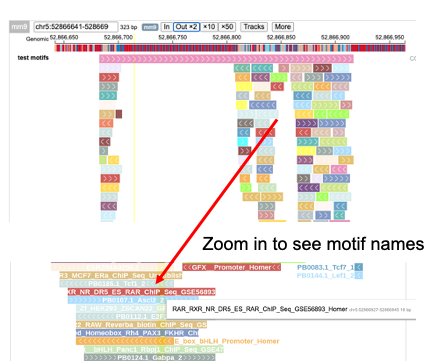
{"type":"bedj","name":"NAME","file":"PATH","stackheight":20,"stackspace":1}
Comments¶
code @ github.