Add gene annotations to CHANGE-seq off-targets table¶
Summary¶
Annotate off-targets using gencode gtf file (e.g., exon, intron, TSS, intergenic).
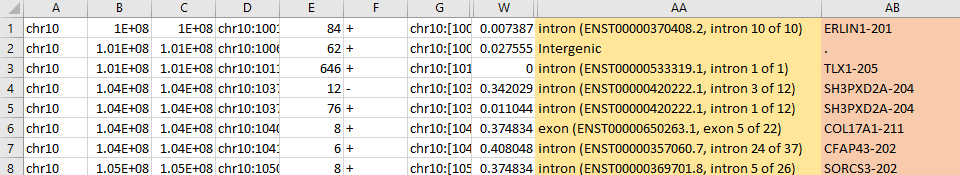
An example output shown below, two addtional columns are added to the original CHANGE-seq table; they are genomic features and gene name (if off-targets occur in exon, intro, TSS, or TTS.)
Usage¶
Step 1
hpcf_interactive -q standard -R "rusage[mem=16000]"
Step 2
module purge
module load python/2.7.13 homer/4.9.1
Step 3
To see the help message:
python /home/yli11/HemTools/bin/add_gene_annotation.py -h
To run an example, takes about 5 min:
python /home/yli11/HemTools/bin/add_gene_annotation.py -f CRL688_CCR5_site_4_identified_matched.txt
output file: CRL688_CCR5_site_4_identified_matched.annot.txt