Upload your bw and bed files to protein paint¶
REF¶
https://docs.google.com/document/d/1e0JVdcf1yQDZst3j77Xeoj_hDN72B6XZ1bo_cAd2rss/edit#
Follow this to use the new track collection version.
Example: https://ppr.stjude.org/?genome=hg19&block=1&tkjsonfile=yli11/rick/test3.json
Summary¶
An easy way to visualizing your data. This program will upload all .bw, .bed, and Peak (newly added, .bedpe and .mango) files to protein paint. Note that protein paint genome browser is only accessible inside stjude network.
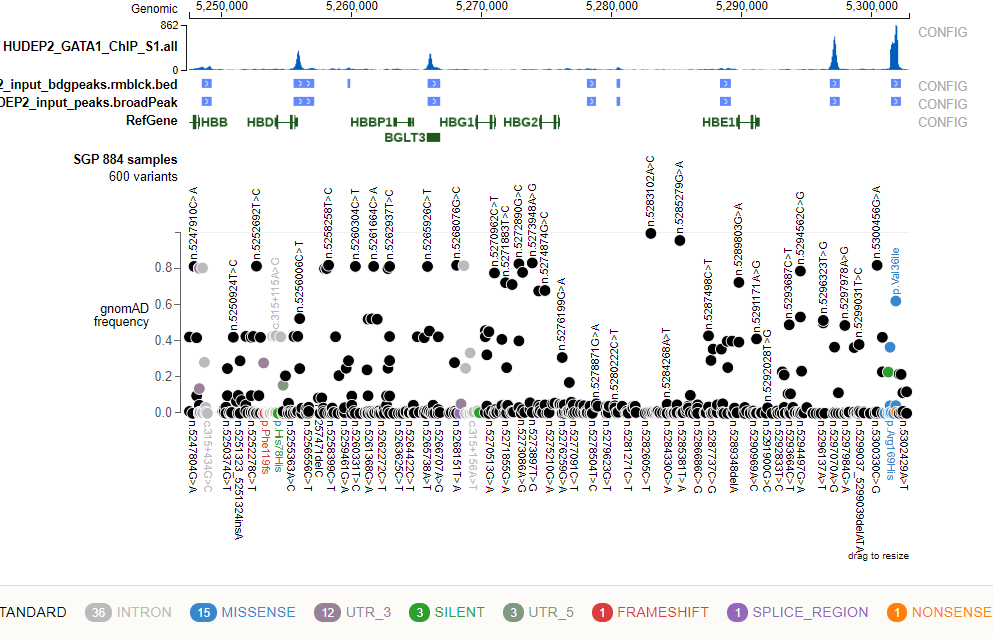
Usage¶
Step 1
module purge
module load python/2.7.13 htslib
Step 2
create_tracks.py -h
usage: create_tracks.py [-h] [-j JID] [-g GENOME] [--current_dir]
optional arguments:
-h, --help show this help message and exit
-j JID, --jid JID a folder name, used to upload tracks (default:
create_tracks_yli11_2019-06-28)
-g GENOME, --genome GENOME
genome version: hg19, hg38, mm9, mm10, hgvirus. (default: hg19)
--current_dir Upload .bed .narrowPeak .broadPeak and .bw files
(default: False)
create_tracks.py --current_dir -g hg19
When finished, it will print out an url, similar like below:
2019-06-28 14:41:43,232 - INFO - upload_bed_bw - connecting to server
2019-06-28 14:41:43,625 - INFO - upload_bed_bw - creating user's dir
2019-06-28 14:41:53,804 - INFO - upload_bed_bw - generating json file
2019-06-28 14:41:56,213 - INFO - upload_bed_bw - transfering file
Please copy the following url to your genome browser. Note that protein paint genome browser is only accessible inside stjude network.
https://ppr.stjude.org/?study=HemPipelines/yli11/create_tracks_yli11_2019-06-283a1f4cad5f47/tracks.json
Add gene track to custom genome¶
download gene annotation gtf file
using lift over bed, to lift over gtf file
using Xin’s tool to convert gtf to bedj
nodejs version >= 2.10
default node mem is 1.5G, increase it to 8G
module load conda3
source activate /home/yli11/.conda/envs/npm
export NODE_OPTIONS=--max-old-space-size=8192
node gtf2bedj.js hg19_20copy.gtf > out.bedjs
sort -k1,1 -k2,2n out.bedjs > hg19_20copy.st.bedj
module load htslib
bgzip hg19_20copy.st.bedj
tabix -p bed hg19_20copy.st.bedj.gz
Put this json:
{
"type":"bedj",
"name":"Ensembl v87 genes",
"file":"yli11/hgcOPT/20copy_data/hg19_20copy.st.bedj.gz",
"stackheight":14,
"stackspace":1
}
automatically generate json file given all files in current_dir¶
cd /home/yli11/dirs/genome_browser/yli11/Jingjing
ppr_json.py -d yli11/Jingjing -o tracks.json -g mm10
Add an existing track to your tracks¶
How to¶
BCL11A motif (hg19)¶
{
"type":"bedj",
"name":"BCL11A(TGACCA)",
"file":"yli11/Chris/TGACCA/BCL11A_TGACCA.bed.bedjs.sorted.gz",
"stackheight":20,
"stackspace":1
}
ZBTB7A motif (hg19)¶
{
"type":"bedj",
"name":"LRF(homer)",
"file":"yli11/motif_tracks/ZBTB7A/LRF-Erythroblasts-ZBTB7A-GSE74977.hg19.fimo.bed.bedjs.sorted.gz",
"stackheight":20,
"stackspace":1
}
GATA1 motif (hg19)¶
{
"type":"bedj",
"name":"GATA1(WGATAR)",
"file":"yli11/Chris/WGATAR/GATA1_WGATAR.bed.bedjs.sorted.gz",
"stackheight":20,
"stackspace":1
}
NFIX motif (hg19)¶
{"type":"bedj","name":"NFIX.hg19.bed.bedjs","file":"HemPipelines/yli11/create_tracks_syi_2021-08-26614c9833fb94/NFIX.hg19.bed.bedjs.sorted.gz","stackheight":10,"stackspace":1}
CTCF motif (hg19)¶
{"type":"bedj","name":"CTCF.hg19.bed.bedjs","file":"HemPipelines/yli11/create_tracks_syi_2021-08-26614c9833fb94/CTCF.hg19.bed.bedjs.sorted.gz","stackheight":10,"stackspace":1}
KLF1 motifs (hg19)¶
{
"type":"bedj",
"name":"KLF1 motifs",
"file":"yli11/CRM_gRNA/KLF_hg19.bed.bedjs.sorted.gz",
"stackheight":20,
"stackspace":1
}
Other motifs (hg19)¶
{
"type":"bedj",
"name":"GATA_Ebox(8-9) motif",
"file":"yli11/CRM_gRNA/complete_per_A_scores/gata_ebox.bed.bedjs.sorted.gz",
"stackheight":20,
"stackspace":1
}
{
"type":"bedj",
"name":"LRF(MA0750.2)",
"file":"yli11/motif_tracks/ZBTB7A/JASPA_MA0750.2_ZBTB7A.hg19.fimo.bed.bedjs.sorted.gz",
"stackheight":20,
"stackspace":1
}
{
"type":"bedj",
"name":"LRF(MA0750.1)",
"file":"yli11/motif_tracks/ZBTB7A/HSAPIENS-ENCODE-MOTIFS-ZBTB7A_KNOWN3_MA0750.1.hg19.fimo.bed.bedjs.sorted.gz",
"stackheight":20,
"stackspace":1
}