ChiCMaxima algorithm review¶
[2]:
file="20copy.called_interactions.tsv"
df = read.table(file,sep="\t",header=T)
head(df)
| ID_Bait | chr_Bait | start_Bait | end_Bait | Bait_name | ID_OE | chr_OE | start_OE | end_OE | OE_name | N | predicted_value | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <int> | <chr> | <int> | <int> | <chr> | <int> | <chr> | <int> | <int> | <chr> | <dbl> | <dbl> | |
| 1 | 32626000 | chr11 | 32626000 | 32626250 | chr11_32625911_32625920_CCDC73 | 307165 | chr11 | 31627500 | 31627750 | chr11_31627500_31627750 | 1.382501 | 1.540992 |
| 2 | 32626000 | chr11 | 32626000 | 32626250 | chr11_32625911_32625920_CCDC73 | 307166 | chr11 | 31627750 | 31628000 | chr11_31627750_31628000 | 1.382501 | 1.540992 |
| 3 | 32626000 | chr11 | 32626000 | 32626250 | chr11_32625911_32625920_CCDC73 | 307167 | chr11 | 31628000 | 31628250 | chr11_31628000_31628250 | 1.382501 | 1.540992 |
| 4 | 32626000 | chr11 | 32626000 | 32626250 | chr11_32625911_32625920_CCDC73 | 307168 | chr11 | 31628250 | 31628500 | chr11_31628250_31628500 | 1.382501 | 1.540992 |
| 5 | 32626000 | chr11 | 32626000 | 32626250 | chr11_32625911_32625920_CCDC73 | 307169 | chr11 | 31628500 | 31628750 | chr11_31628500_31628750 | 1.382501 | 1.540992 |
| 6 | 32626000 | chr11 | 32626000 | 32626250 | chr11_32625911_32625920_CCDC73 | 307170 | chr11 | 31628750 | 31629000 | chr11_31628750_31629000 | 1.382501 | 1.540992 |
[4]:
df2 = subset(df,Bait_name=="chr1_235493350_235493351_GGPS1")
[5]:
head(df2)
| ID_Bait | chr_Bait | start_Bait | end_Bait | Bait_name | ID_OE | chr_OE | start_OE | end_OE | OE_name | N | predicted_value | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <int> | <chr> | <int> | <int> | <chr> | <int> | <chr> | <int> | <int> | <chr> | <dbl> | <dbl> | |
| 54355 | 235493500 | chr1 | 235493500 | 235493750 | chr1_235493350_235493351_GGPS1 | 167002 | chr1 | 234493250 | 234493500 | chr1_234493250_234493500 | 1.612917 | 1.212972 |
| 54356 | 235493500 | chr1 | 235493500 | 235493750 | chr1_235493350_235493351_GGPS1 | 167003 | chr1 | 234493500 | 234493750 | chr1_234493500_234493750 | 2.172501 | 1.212972 |
| 54357 | 235493500 | chr1 | 235493500 | 235493750 | chr1_235493350_235493351_GGPS1 | 167004 | chr1 | 234493750 | 234494000 | chr1_234493750_234494000 | 2.304168 | 1.212972 |
| 54358 | 235493500 | chr1 | 235493500 | 235493750 | chr1_235493350_235493351_GGPS1 | 167005 | chr1 | 234494000 | 234494250 | chr1_234494000_234494250 | 2.488501 | 1.212972 |
| 54359 | 235493500 | chr1 | 235493500 | 235493750 | chr1_235493350_235493351_GGPS1 | 167006 | chr1 | 234494250 | 234494500 | chr1_234494250_234494500 | 2.488501 | 1.212972 |
| 54360 | 235493500 | chr1 | 235493500 | 235493750 | chr1_235493350_235493351_GGPS1 | 167007 | chr1 | 234494500 | 234494750 | chr1_234494500_234494750 | 2.765001 | 1.212972 |
[6]:
plot(df2$N)
[10]:
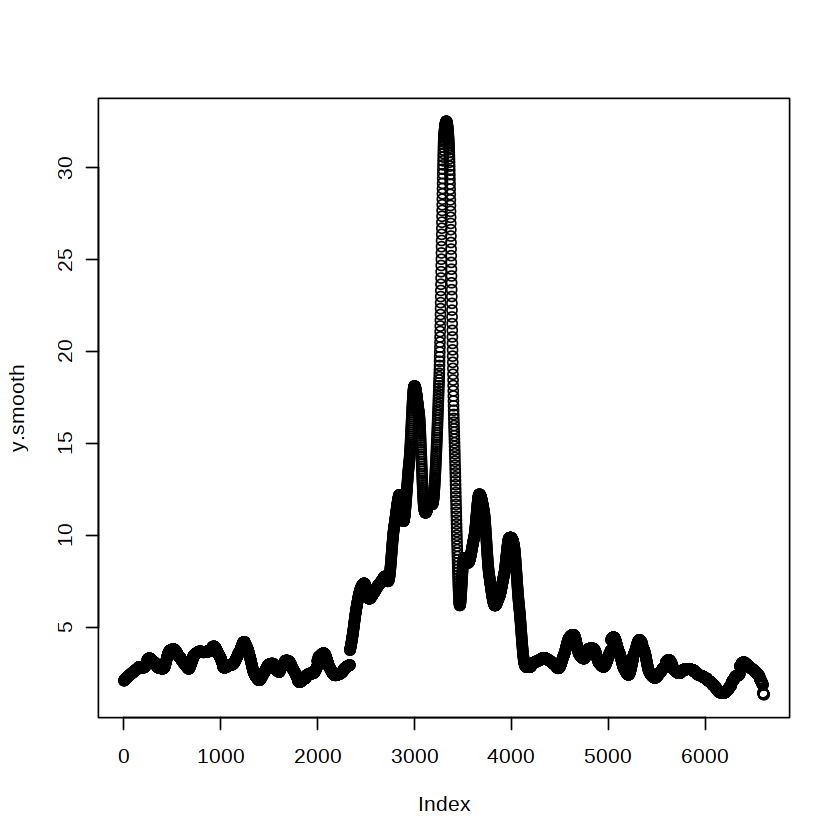
y=df2[,"N"]
x=df2[,"start_OE"]
n <- length(y)
span=0.05
y.smooth <- loess(y ~ x, span=span)$fitted
[11]:
plot(y)
[12]:
plot(y.smooth)
[14]:
library(zoo)
Attaching package: ‘zoo’
The following objects are masked from ‘package:base’:
as.Date, as.Date.numeric
[16]:
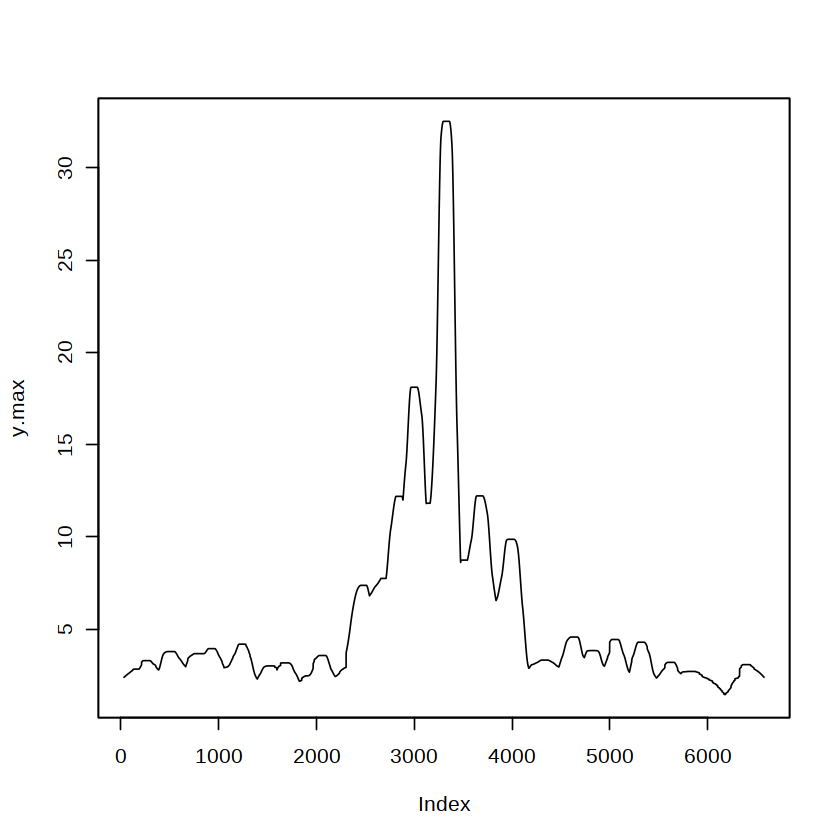
w=30
y.max <- rollapply(zoo(y.smooth), 2*w+1,max,align="center")
[17]:
plot(y.max)
[18]:
delta <- y.max - y.smooth[-c(1:w, n+1-1:w)]
[19]:
plot(delta)
[20]:
d=0
i.max <- which(delta <= d) + w
[29]:
?rollapply
| rollapply {zoo} | R Documentation |
Apply Rolling Functions
Description
A generic function for applying a function to rolling margins of an array.
Usage
rollapply(data, ...)
## S3 method for class 'ts'
rollapply(data, ...)
## S3 method for class 'zoo'
rollapply(data, width, FUN, ..., by = 1, by.column = TRUE,
fill = if (na.pad) NA, na.pad = FALSE, partial = FALSE,
align = c("center", "left", "right"), coredata = TRUE)
## Default S3 method:
rollapply(data, ...)
rollapplyr(..., align = "right")
Arguments
data |
the data to be used (representing a series of observations). |
width |
numeric vector or list. In the simplest case this is an integer
specifying the window width (in numbers of observations) which is aligned
to the original sample according to the |
FUN |
the function to be applied. |
... |
optional arguments to |
by |
calculate FUN at every |
by.column |
logical. If |
fill |
a three-component vector or list (recycled otherwise) providing
filling values at the left/within/to the right of the data range.
See the |
na.pad |
deprecated. Use |
partial |
logical or numeric. If |
align |
specifyies whether the index of the result
should be left- or right-aligned or centered (default) compared
to the rolling window of observations. This argument is only used if
|
coredata |
logical. Should only the |
Details
If width is a plain numeric vector its elements are regarded as widths
to be interpreted in conjunction with align whereas if width is a list
its components are regarded as offsets. In the above cases if the length of
width is 1 then width is recycled for every by-th point.
If width is a list its components represent integer offsets such that
the i-th component of the list refers to time points at positions
i + width[[i]]. If any of these points are below 1 or above the
length of index(data) then FUN is not evaluated for that
point unless partial = TRUE and in that case only the valid
points are passed.
The rolling function can also be applied to partial windows by setting partial = TRUE
For example, if width = 3, align = "right" then for the first point
just that point is passed to FUN since the two points to its
left are out of range. For the same example, if partial = FALSE then FUN is not
invoked at all for the first two points. If partial is a numeric then it
specifies the minimum number of offsets that must be within range. Negative
partial is interpreted as FALSE.
If width is a scalar then partial = TRUE and fill = NA are
mutually exclusive but if offsets are specified for the width and 0 is not
among the offsets then the output will be shorter than the input even
if partial = TRUE is specified. In that case it may still be useful
to specify fill in addition to partial.
If FUN is mean, max or median and by.column is
TRUE and width is a plain scalar and there are no other arguments
then special purpose code is used to enhance performance.
Also in the case of mean such special purpose code is only invoked if the
data argument has no NA values.
See rollmean, rollmax and rollmedian
for more details.
Currently, there are methods for "zoo" and "ts" series
and "default" method for ordinary vectors and matrices.
rollapplyr is a wrapper around rollapply that uses a default
of align = "right".
If data is of length 0, data is returned unmodified.
Value
A object of the same class as data with the results of the rolling function.
See Also
rollmean
Examples
suppressWarnings(RNGversion("3.5.0"))
set.seed(1)
## rolling mean
z <- zoo(11:15, as.Date(31:35))
rollapply(z, 2, mean)
## non-overlapping means
z2 <- zoo(rnorm(6))
rollapply(z2, 3, mean, by = 3) # means of nonoverlapping groups of 3
aggregate(z2, c(3,3,3,6,6,6), mean) # same
## optimized vs. customized versions
rollapply(z2, 3, mean) # uses rollmean which is optimized for mean
rollmean(z2, 3) # same
rollapply(z2, 3, (mean)) # does not use rollmean
## rolling regression:
## set up multivariate zoo series with
## number of UK driver deaths and lags 1 and 12
seat <- as.zoo(log(UKDriverDeaths))
time(seat) <- as.yearmon(time(seat))
seat <- merge(y = seat, y1 = lag(seat, k = -1),
y12 = lag(seat, k = -12), all = FALSE)
## run a rolling regression with a 3-year time window
## (similar to a SARIMA(1,0,0)(1,0,0)_12 fitted by OLS)
rr <- rollapply(seat, width = 36,
FUN = function(z) coef(lm(y ~ y1 + y12, data = as.data.frame(z))),
by.column = FALSE, align = "right")
## plot the changes in coefficients
## showing the shifts after the oil crisis in Oct 1973
## and after the seatbelt legislation change in Jan 1983
plot(rr)
## rolling mean by time window (e.g., 3 days) rather than
## by number of observations (e.g., when these are unequally spaced):
#
## - test data
tt <- as.Date("2000-01-01") + c(1, 2, 5, 6, 7, 8, 10)
z <- zoo(seq_along(tt), tt)
## - fill it out to a daily series, zm, using NAs
## using a zero width zoo series g on a grid
g <- zoo(, seq(start(z), end(z), "day"))
zm <- merge(z, g)
## - 3-day rolling mean
rollapply(zm, 3, mean, na.rm = TRUE, fill = NA)
##
## - without expansion to regular grid: find interval widths
## that encompass the previous 3 days for each Date
w <- seq_along(tt) - findInterval(tt - 3, tt)
## a solution to computing the widths 'w' that is easier to read but slower
## w <- sapply(tt, function(x) sum(tt >= x - 2 & tt <= x))
##
## - rolling sum from 3-day windows
## without vs. with expansion to regular grid
rollapplyr(z, w, sum)
rollapplyr(zm, 3, sum, partial = TRUE, na.rm = TRUE)
## rolling weekly sums (with some missing dates)
z <- zoo(1:11, as.Date("2016-03-09") + c(0:7, 9:10, 12))
weeksum <- function(z) sum(z[time(z) > max(time(z)) - 7])
zs <- rollapplyr(z, 7, weeksum, fill = NA, coredata = FALSE)
merge(value = z, weeksum = zs)
## replicate cumsum with either 'partial' or vector width 'k'
cumsum(1:10)
rollapplyr(1:10, 10, sum, partial = TRUE)
rollapplyr(1:10, 1:10, sum)
## different values of rule argument
z <- zoo(c(NA, NA, 2, 3, 4, 5, NA))
rollapply(z, 3, sum, na.rm = TRUE)
rollapply(z, 3, sum, na.rm = TRUE, fill = NULL)
rollapply(z, 3, sum, na.rm = TRUE, fill = NA)
rollapply(z, 3, sum, na.rm = TRUE, partial = TRUE)
# this will exclude time points 1 and 2
# It corresponds to align = "right", width = 3
rollapply(zoo(1:8), list(seq(-2, 0)), sum)
# but this will include points 1 and 2
rollapply(zoo(1:8), list(seq(-2, 0)), sum, partial = 1)
rollapply(zoo(1:8), list(seq(-2, 0)), sum, partial = 0)
# so will this
rollapply(zoo(1:8), list(seq(-2, 0)), sum, fill = NA)
# by = 3, align = "right"
L <- rep(list(NULL), 8)
L[seq(3, 8, 3)] <- list(seq(-2, 0))
str(L)
rollapply(zoo(1:8), L, sum)
rollapply(zoo(1:8), list(0:2), sum, fill = 1:3)
rollapply(zoo(1:8), list(0:2), sum, fill = 3)
L2 <- rep(list(-(2:0)), 10)
L2[5] <- list(NULL)
str(L2)
rollapply(zoo(1:10), L2, sum, fill = "extend")
rollapply(zoo(1:10), L2, sum, fill = list("extend", NULL))
rollapply(zoo(1:10), L2, sum, fill = list("extend", NA))
rollapply(zoo(1:10), L2, sum, fill = NA)
rollapply(zoo(1:10), L2, sum, fill = 1:3)
rollapply(zoo(1:10), L2, sum, partial = TRUE)
rollapply(zoo(1:10), L2, sum, partial = TRUE, fill = 99)
rollapply(zoo(1:10), list(-1), sum, partial = 0)
rollapply(zoo(1:10), list(-1), sum, partial = TRUE)
rollapply(zoo(cbind(a = 1:6, b = 11:16)), 3, rowSums, by.column = FALSE)
# these two are the same
rollapply(zoo(cbind(a = 1:6, b = 11:16)), 3, sum)
rollapply(zoo(cbind(a = 1:6, b = 11:16)), 3, colSums, by.column = FALSE)
# these two are the same
rollapply(zoo(1:6), 2, sum, by = 2, align = "right")
aggregate(zoo(1:6), c(2, 2, 4, 4, 6, 6), sum)
# these two are the same
rollapply(zoo(1:3), list(-1), c)
lag(zoo(1:3), -1)
# these two are the same
rollapply(zoo(1:3), list(1), c)
lag(zoo(1:3))
# these two are the same
rollapply(zoo(1:5), list(c(-1, 0, 1)), sum)
rollapply(zoo(1:5), 3, sum)
# these two are the same
rollapply(zoo(1:5), list(0:2), sum)
rollapply(zoo(1:5), 3, sum, align = "left")
# these two are the same
rollapply(zoo(1:5), list(-(2:0)), sum)
rollapply(zoo(1:5), 3, sum, align = "right")
# these two are the same
rollapply(zoo(1:6), list(NULL, NULL, -(2:0)), sum)
rollapply(zoo(1:6), 3, sum, by = 3, align = "right")
# these two are the same
rollapply(zoo(1:5), list(c(-1, 1)), sum)
rollapply(zoo(1:5), 3, function(x) sum(x[-2]))
# these two are the same
rollapply(1:5, 3, rev)
embed(1:5, 3)
# these four are the same
x <- 1:6
rollapply(c(0, 0, x), 3, sum, align = "right") - x
rollapply(x, 3, sum, partial = TRUE, align = "right") - x
rollapply(x, 3, function(x) sum(x[-3]), partial = TRUE, align = "right")
rollapply(x, list(-(2:1)), sum, partial = 0)
# same as Matlab's buffer(x, n, p) for valid non-negative p
# See http://www.mathworks.com/help/toolbox/signal/buffer.html
x <- 1:30; n <- 7; p <- 3
t(rollapply(c(rep(0, p), x, rep(0, n-p)), n, by = n-p, c))
# these three are the same
y <- 10 * seq(8); k <- 4; d <- 2
# 1
# from http://ucfagls.wordpress.com/2011/06/14/embedding-a-time-series-with-time-delay-in-r-part-ii/
Embed <- function(x, m, d = 1, indices = FALSE, as.embed = TRUE) {
n <- length(x) - (m-1)*d
X <- seq_along(x)
if(n <= 0)
stop("Insufficient observations for the requested embedding")
out <- matrix(rep(X[seq_len(n)], m), ncol = m)
out[,-1] <- out[,-1, drop = FALSE] +
rep(seq_len(m - 1) * d, each = nrow(out))
if(as.embed)
out <- out[, rev(seq_len(ncol(out)))]
if(!indices)
out <- matrix(x[out], ncol = m)
out
}
Embed(y, k, d)
# 2
rollapply(y, list(-d * seq(0, k-1)), c)
# 3
rollapply(y, d*k-1, function(x) x[d * seq(k-1, 0) + 1])
## mimic convolve() using rollapplyr()
A <- 1:4
B <- 5:8
## convolve(..., type = "open")
cross <- function(x) x
rollapplyr(c(A, 0*B[-1]), length(B), cross, partial = TRUE)
convolve(A, B, type = "open")
# convolve(..., type = "filter")
rollapplyr(A, length(B), cross)
convolve(A, B, type = "filter")
# weighted sum including partials near ends, keeping
## alignment with wts correct
points <- zoo(cbind(lon = c(11.8300715, 11.8296697,
11.8268708, 11.8267236, 11.8249612, 11.8251062),
lat = c(48.1099048, 48.10884, 48.1067431, 48.1066077,
48.1037673, 48.103318),
dist = c(46.8463805878941, 33.4921440879536, 10.6101735030534,
18.6085009578724, 6.97253109610173, 9.8912817449265)))
mysmooth <- function(z, wts = c(0.3, 0.4, 0.3)) {
notna <- !is.na(z)
sum(z[notna] * wts[notna]) / sum(wts[notna])
}
points2 <- points
points2[, 1:2] <- rollapply(rbind(NA, coredata(points)[, 1:2], NA), 3, mysmooth)
points2
[28]:
plot(zoo(y.smooth))
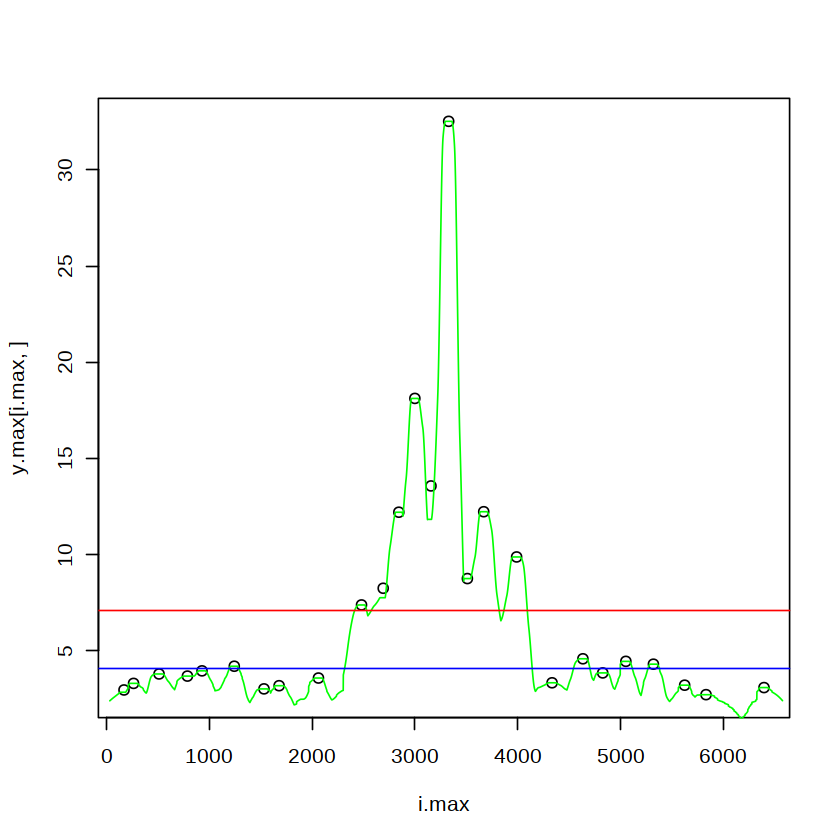
[38]:
plot(x=i.max,y=y.max[i.max,])
lines(y.max,col="green")
abline(h=median(y.max[i.max,]),col="blue")
abline(h=mean(y.max[i.max,]),col="red")
[37]:
?abline
| abline {graphics} | R Documentation |
Add Straight Lines to a Plot
Description
This function adds one or more straight lines through the current plot.
Usage
abline(a = NULL, b = NULL, h = NULL, v = NULL, reg = NULL,
coef = NULL, untf = FALSE, ...)
Arguments
a, b |
the intercept and slope, single values. |
untf |
logical asking whether to untransform. See ‘Details’. |
h |
the y-value(s) for horizontal line(s). |
v |
the x-value(s) for vertical line(s). |
coef |
a vector of length two giving the intercept and slope. |
reg |
an object with a |
... |
graphical parameters such as
|
Details
Typical usages are
abline(a, b, untf = FALSE, \dots) abline(h =, untf = FALSE, \dots) abline(v =, untf = FALSE, \dots) abline(coef =, untf = FALSE, \dots) abline(reg =, untf = FALSE, \dots)
The first form specifies the line in intercept/slope form
(alternatively a can be specified on its own and is taken to
contain the slope and intercept in vector form).
The h= and v= forms draw horizontal and vertical lines
at the specified coordinates.
The coef form specifies the line by a vector containing the
slope and intercept.
reg is a regression object with a coef method.
If this returns a vector of length 1 then the value is taken to be the
slope of a line through the origin, otherwise, the first 2 values are
taken to be the intercept and slope.
If untf is true, and one or both axes are log-transformed, then
a curve is drawn corresponding to a line in original coordinates,
otherwise a line is drawn in the transformed coordinate system. The
h and v parameters always refer to original coordinates.
The graphical parameters col, lty and lwd
can be specified; see par for details. For the
h= and v= usages they can be vectors of length greater
than one, recycled as necessary.
Specifying an xpd argument for clipping overrides
the global par("xpd") setting used otherwise.
References
Becker, R. A., Chambers, J. M. and Wilks, A. R. (1988) The New S Language. Wadsworth & Brooks/Cole.
Murrell, P. (2005) R Graphics. Chapman & Hall/CRC Press.
See Also
lines and segments for connected and
arbitrary lines given by their endpoints.
par.
Examples
## Setup up coordinate system (with x == y aspect ratio): plot(c(-2,3), c(-1,5), type = "n", xlab = "x", ylab = "y", asp = 1) ## the x- and y-axis, and an integer grid abline(h = 0, v = 0, col = "gray60") text(1,0, "abline( h = 0 )", col = "gray60", adj = c(0, -.1)) abline(h = -1:5, v = -2:3, col = "lightgray", lty = 3) abline(a = 1, b = 2, col = 2) text(1,3, "abline( 1, 2 )", col = 2, adj = c(-.1, -.1)) ## Simple Regression Lines: require(stats) sale5 <- c(6, 4, 9, 7, 6, 12, 8, 10, 9, 13) plot(sale5) abline(lsfit(1:10, sale5)) abline(lsfit(1:10, sale5, intercept = FALSE), col = 4) # less fitting z <- lm(dist ~ speed, data = cars) plot(cars) abline(z) # equivalent to abline(reg = z) or abline(coef = coef(z)) ## trivial intercept model abline(mC <- lm(dist ~ 1, data = cars)) ## the same as abline(a = coef(mC), b = 0, col = "blue")