Analysis pipeline for change-seq randomized assay¶
Library Structure¶
Input¶
8-columns tsv file (no header). The first 3 columns are R1, R2 and sample name. Save this file’s name as input.tsv. The pipeline uses input.tsv for downstream analysis.
The rest columns are meta information, including 5` adapter sequence, 3` adapter sequence, gRNA name, whether it is a treatment nuclease or control control. The last column is the gRNA sequence (include PAM).
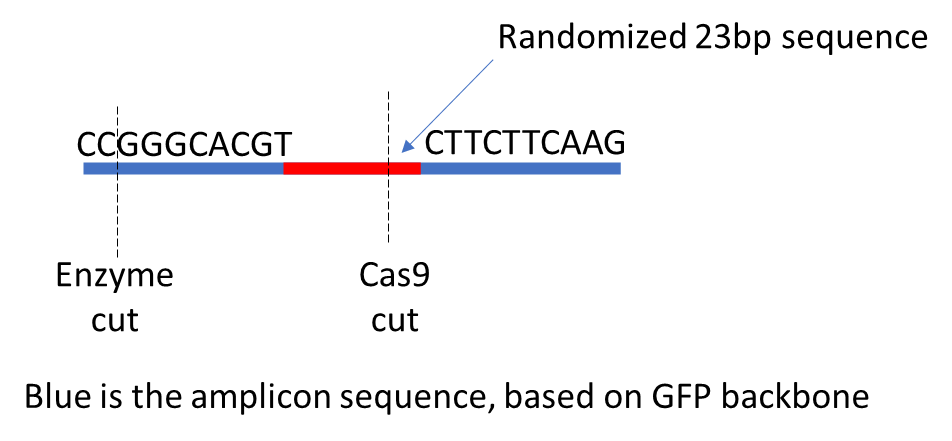
ABC_R1_001.fastq.gz ABC_R2_001.fastq.gz ABC CCGGGCACGT CTTCTTCAAG gRNA_name nuclease GTCCCTAGTGGCCCCACTGTGGG
Orange_S2_R1_001.fastq.gz Orange_S2_R2_001.fastq.gz Orange CCGGGCACGT CTTCTTCAAG gRNA_name control GTCCCTAGTGGCCCCACTGTGGG
gRNA sequence reference:
AAVS1_s2 GTCCCTAGTGGCCCCACTGTGGG
AAVS1_s11 GGTGAGGGAGGAGAGATGCCCGG
AAVS1_s14 GGGGCCACTAGGGACAGGATTGG
CTLA4_s9 GGACTGAGGGCCATGGACACGGG
CCR5_s8 GGACAGTAAGAAGGAAAAACAGG
TRAC_s1 GTCAGGGTTCTGGATATCTGTGG
LAG3_s9 GAAGGCTGAGATCCTGGAGGGGG
CXCR4_s8 GTCCCCTGAGCCCATTTCCTCGG
Usage¶
# copy your fastq file to the working dir
hpcf_interactive # login to a compute node
module load python/2.7.13
run_lsf.py -f input.tsv -u /home/yli11/Tools/changeseq_randomized/version6_combined.lsf