Motif notes¶
FIMO¶
5-mer exact match p-value cutoff: 0.001 10-mer allows two mismatches: 1e-4 (default cutoff)
AGATAA |
0.000352 |
AGATAG |
0.00028900000000000003 |
CCAAT |
0.00105 |
TGACCA |
0.00023700000000000001 |
TGATAA |
0.000352 |
TGATAG |
0.00028900000000000003 |
p_value¶
https://groups.google.com/forum/#!topic/meme-suite/xD2Z3EUw2BQ
the default 1e-4 p-value is not likely to report any matches for motif length=5,6
Motif mapping bed file is generated using FIMO. For motif length <= 7, p-value cutoff is 1e-3. For motif length >=8, default cutoff 1e-4 is used.
motif length |
p_value cutoff |
5-7 |
1e-3 |
8+ |
1e-4 (default) |
bedtools¶
http://quinlanlab.org/tutorials/bedtools/bedtools.html#intersecting-multiple-files-at-once.
CTCF motif¶
CTCF motif is a long conserved motif in mammalian lineage.
This paper in 2012 divided CTCF motif into M1 + M2, a new 2020 genome biology paper divided it into M1-M4. Visually, they are the same.
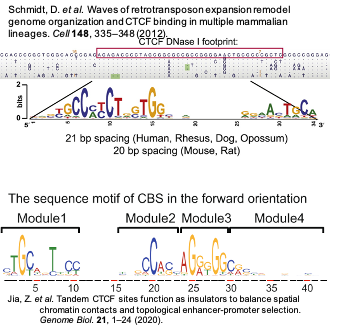
You can find the pwm here: http://insulatordb.uthsc.edu/download/CTCFBSDB_PWM.mat
http://genesdev.cshlp.org/content/30/8/881.full.pdf
https://www.nature.com/articles/s41467-018-06664-6
https://www.biorxiv.org/content/10.1101/2020.07.02.185389v1.full.pdf