Plot motif position density on peaks¶
usage: plot_homer_motif_peak.py [-h] -f BED_FILE [-g GENOME]
[--flanking_size FLANKING_SIZE]
[--bin_size BIN_SIZE]
optional arguments:
-h, --help show this help message and exit
-f BED_FILE, --bed_file BED_FILE
bed or peak file (default: None)
Genome Info:
-g GENOME, --genome GENOME
genome version: hg19, mm10, mm9 (default: hg19)
Plot Info:
--flanking_size FLANKING_SIZE
extend this size to left and right (default: 1000)
--bin_size BIN_SIZE bin size for plot density, use minimal 10 maximal 50,
motif size can affect the figure a little bit, if you
want to make the peak stronger, change this parameter
(default: 20)
Example¶
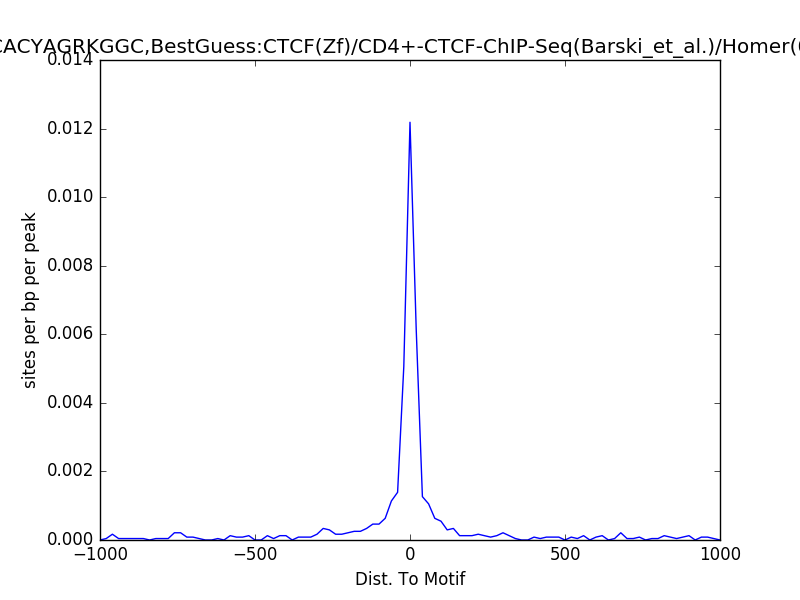
Usage¶
Suppose you have done homer motif discovery, then go to your motif discovery result folder, in this folder you should see two folders named homerResults and knownResults.
Step 0: Load python version 2.7.13.
module load python/2.7.13
module load homer
Step 1: Run
plot_homer_motif_peak.py -f peak_file.bed
Output¶
The output folder is homer_all_motifs; it contains png files for every motif.
Comments¶
code @ github.