Plot correlation scatter plots¶
usage: scatter_density.py [-h] -f F [-s S] -x X -y Y [--index INDEX]
[--regression] [--diagnal_line] [--lowess]
[-bc BACKGROUND_DOTS_COLOR] [-hc HIGHLIGHT_COLOR]
[--highlight HIGHLIGHT] [-o OUTPUT]
optional arguments:
-h, --help show this help message and exit
-f F data table (default: None)
-s S sep (default: )
-x X column name for x axis (default: None)
-y Y column name for y axis (default: None)
--index INDEX index name for index (default: None)
--regression by default it is a dignal line (default: False)
--diagnal_line force to draw a dignal line (default: False)
--lowess fit a curve (default: False)
-bc BACKGROUND_DOTS_COLOR, --background_dots_color BACKGROUND_DOTS_COLOR
background_dots_color (default: #0000ff)
-hc HIGHLIGHT_COLOR, --highlight_color HIGHLIGHT_COLOR
highlight_color (default: #ff1500)
--highlight HIGHLIGHT
column name for y axis, sep by comma (default: None)
-o OUTPUT, --output OUTPUT
output file name (default:
yli11_2020-09-22.correlation_scatter_density.png)
Summary¶
This script can be used to calculate sample correlation. Values are log-transformed, e.g., log2(x+1). Addtionally, you can also highlight some points, for example, to show some differences.
Input¶
Input can be tsv or csv. For tsv use -s "\t", for csv use -s ,
Geneid Chr Start End Strand Length Banana Orange
asd1 chr1 3513707 3514076 + 370 800 22
b chr1 3538168 3538438 + 271 24 16
a chr1 3970540 3970785 + 246 16 6
b chr1 4059120 4059436 + 317 44 12
a chr1 4388977 4389294 + 318 22 11
b chr1 4561768 4562101 + 334 31 11
a chr1 4760133 4760340 + 208 23 9
b chr1 5073062 5073299 + 238 36 20
The above example is a read count distribution for two chip-seq replicates.
The aim is to see the correlation between Banana and Orange, use -x Banana -y Orange to plot Banana column as the X-axis and Orange column as the Y-axis.
Usage¶
Sample correlation usage¶
hpcf_interactive -q standard -R "rusage[mem=10000]"
module load conda3
source activate /home/yli11/.conda/envs/py2/
scatter_density.py -f input.tsv -s "\t" -x Banana -y Orange
Point highlight usage¶
hpcf_interactive -q standard -R "rusage[mem=10000]"
module load conda3
source activate /home/yli11/.conda/envs/py2/
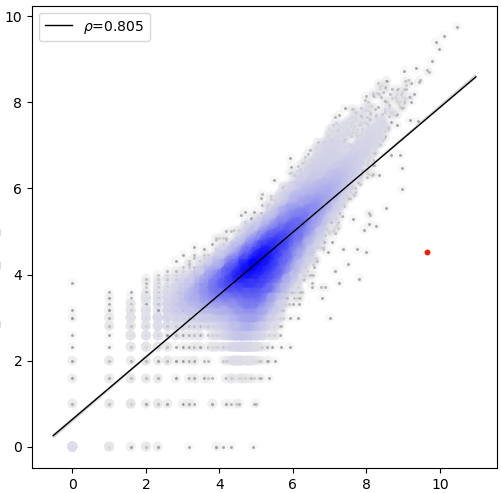
scatter_density.py -f input.tsv -s "\t" -x Banana -y Orange --index Geneid --highlight asd1 --regression
--regression is to add regression line. For sample correlation plot, regression line is on, for differential plot, the default is off.
If you have multiple points to highlight, use --highlight asd1,another_name,another_name2. These names should match the index name, which is defined using --index Geneid
Output¶
sample correlation¶
The value shown on the upper left corner is pearson correlation coefficicient.
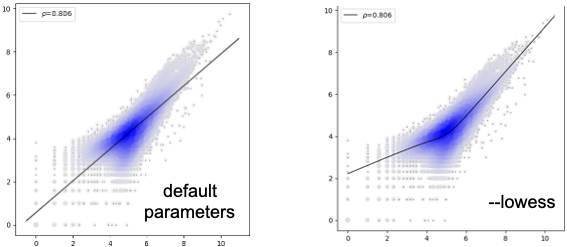
differential analysis highlight¶
The value shown on the upper left corner is pearson correlation coefficicient.