Test differences in number of interactions¶
usage: overlap_bedpe.py [-h] -p POS_BED -n NEG_BED -f BEDPE [-o OUTPUT]
optional arguments:
-h, --help show this help message and exit
-p POS_BED, --pos_bed POS_BED
positive bed file, chr, start, end, etc. (default:
None)
-n NEG_BED, --neg_bed NEG_BED
negative bed file, chr, start, end, etc. (default:
None)
-f BEDPE, --bedpe BEDPE
bedpe, chr, start, end, chr, start, end, etc.
(default: None)
-o OUTPUT, --output OUTPUT
Summary¶
Given a positive and a negative bed file, get number of interactions for each region in the two bed files and apply welch’s t-test to get the significance.
Example¶
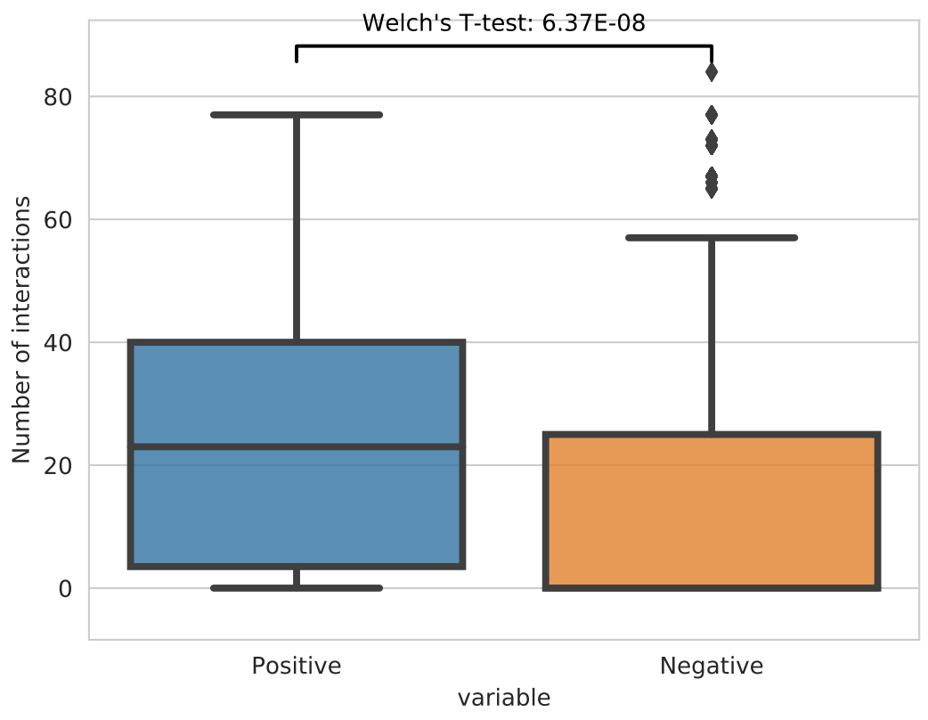
Input¶
two bed file and a bedpe file.
Example of bedpe file: only the first 6 columns are used. You might want to first extract significant interactions then perform our analysis.
chr1 62099 63943 chr1 105955 107671 1 0.90552424887951
chr1 105955 107671 chr1 235358 237015 1 0.051453442758473
chr1 712256 716178 chr1 753657 755899 1 0.90552424887951
chr1 712256 716178 chr1 761417 764406 3 0.34819012369507
chr1 712256 716178 chr1 766795 769182 2 0.660667234099137
Output¶
output.pdf
You can also specify the output file name.
Usage¶
hpcf_interactive
source activate /home/yli11/.conda/envs/py2/
cd /home/yli11/test/xxx/
overlap_bedpe.py -p pos.bed -n neg.bed -f Hudep2_D0_H3K27AC_HiChIP_FS.interactions.all.mango