Identify direct targets and co-binding factors¶
usage: tf_target_finder.py [-h] [-j JID] -q QUERY_BED -exp DEG_TSV
--query_motif QUERY_MOTIF [-tss TSS_BED]
[-epi EPI_BED] [--LFC_col_name LFC_COL_NAME]
[--FDR_col_name FDR_COL_NAME]
[--LFC_cutoff LFC_CUTOFF] [--FDR_cutoff FDR_CUTOFF]
[-d1 D1] [-d2 D2] [-d3 D3] [-d4 D4] [-d5 D5]
[-d6 D6] [--motif_database MOTIF_DATABASE]
[--motif_list MOTIF_LIST] [--peak_list PEAK_LIST]
[--assign_targets_addon_parameters ASSIGN_TARGETS_ADDON_PARAMETERS]
[--label LABEL]
optional arguments:
-h, --help show this help message and exit
-j JID, --jid JID enter a job ID, which is used to make a new directory.
Every output will be moved into this folder. (default:
tf_target_finder_yli11_2020-04-22)
-q QUERY_BED, --query_bed QUERY_BED
3 column bed file, additional columns are OK, but will
be ignored (default: None)
-exp DEG_TSV, --deg_tsv DEG_TSV
any number of columns, first column should be gene
name, first row should be column names. should contain
FDR and LFC. (default: None)
--query_motif QUERY_MOTIF
query_motif pwm file (default: None)
-tss TSS_BED, --tss_bed TSS_BED
4 column bed file, the 4th column should be gene name,
should match to the gene name in DEG file (if
supplied). Additional columns are OK, but will be
ignored (default: /home/yli11/Data/Mouse/mm9/annotatio
ns/mm9.ensembl_v67.TSS.gene_name.bed)
-epi EPI_BED, --epi_bed EPI_BED
5 column bed file, the 4th column should be gene name,
should match to the gene name in DEG file and TSS
annotation(if supplied). The 5th column should be
score (optional). Additional columns are OK, but will
be ignored (default: /home/yli11/Tools/TF_target_finde
r/data/HPC7.mm9.captureC.bed)
--LFC_col_name LFC_COL_NAME
LFC_col_name (default: logFC)
--FDR_col_name FDR_COL_NAME
FDR_col_name (default: adj.P.Val)
--LFC_cutoff LFC_CUTOFF
LFC cutoff (default: 1)
--FDR_cutoff FDR_CUTOFF
FDR cutoff (default: 0.05)
-d1 D1 extend query bed for intersection (default: 0)
-d2 D2 extending tss for intersection (default: 5000)
-d3 D3 extending epi for intersection (default: 0)
-d4 D4 for motif scanning: extend search on the flank
sequences (default: 100)
-d5 D5 distance cutoff for peak overlap, used for co-binding
test (default: 500)
-d6 D6 distance cutoff for motif overlap, used for co-binding
test (default: 100)
--motif_database MOTIF_DATABASE
motif meme file (default:
/home/yli11/Data/Motif_database/Mouse/mouse_TF.meme)
--motif_list MOTIF_LIST
motif_list (default: /home/yli11/HemTools/share/misc/T
F_target_finder/motif.list)
--peak_list PEAK_LIST
peak_list (default: /home/yli11/HemTools/share/misc/TF
_target_finder/peak.list)
--assign_targets_addon_parameters ASSIGN_TARGETS_ADDON_PARAMETERS
any addon parameters (default: )
--label LABEL give a name for your TF (i.e., query) (default:
target_finder)
Summary¶
A common down-stream analysis of ChIP-seq peaks (or more generally, a set of cis-regulatory elements) is to find their target genes. However, assigning distal regulatory elements to their correct target genes is not an easy problem. Systematic comparison of several target gene assignment algorithms based on real promoter capture-C or HiC has found that the best-performing method is only modestly better than a baseline distance method for most benchmark datasets, suggesting that the most confident assignment should be still based on real experiments.
Therefore, our TF_target_finder pipeline uses promoter-enhancer interactions from promoter capture-C or HiC datasets and outputs a list of high-confidence assignments using differentially expressed genes from WT.vs.KO datasets.
Workflow¶
Target genes were assigned not only based on nearest TSS but also based on promoter capture-C, which were then filtered out using an associated RNA-seq experiments (e.g., the knockout of query TF, WT.vs.KO) in which we assume the query TF regulates the differentially expressed genes.
The output of peaks with assigned targets will be used to find co-binding factors given co-factor peaks and motifs.
An overall workflow is shown below.
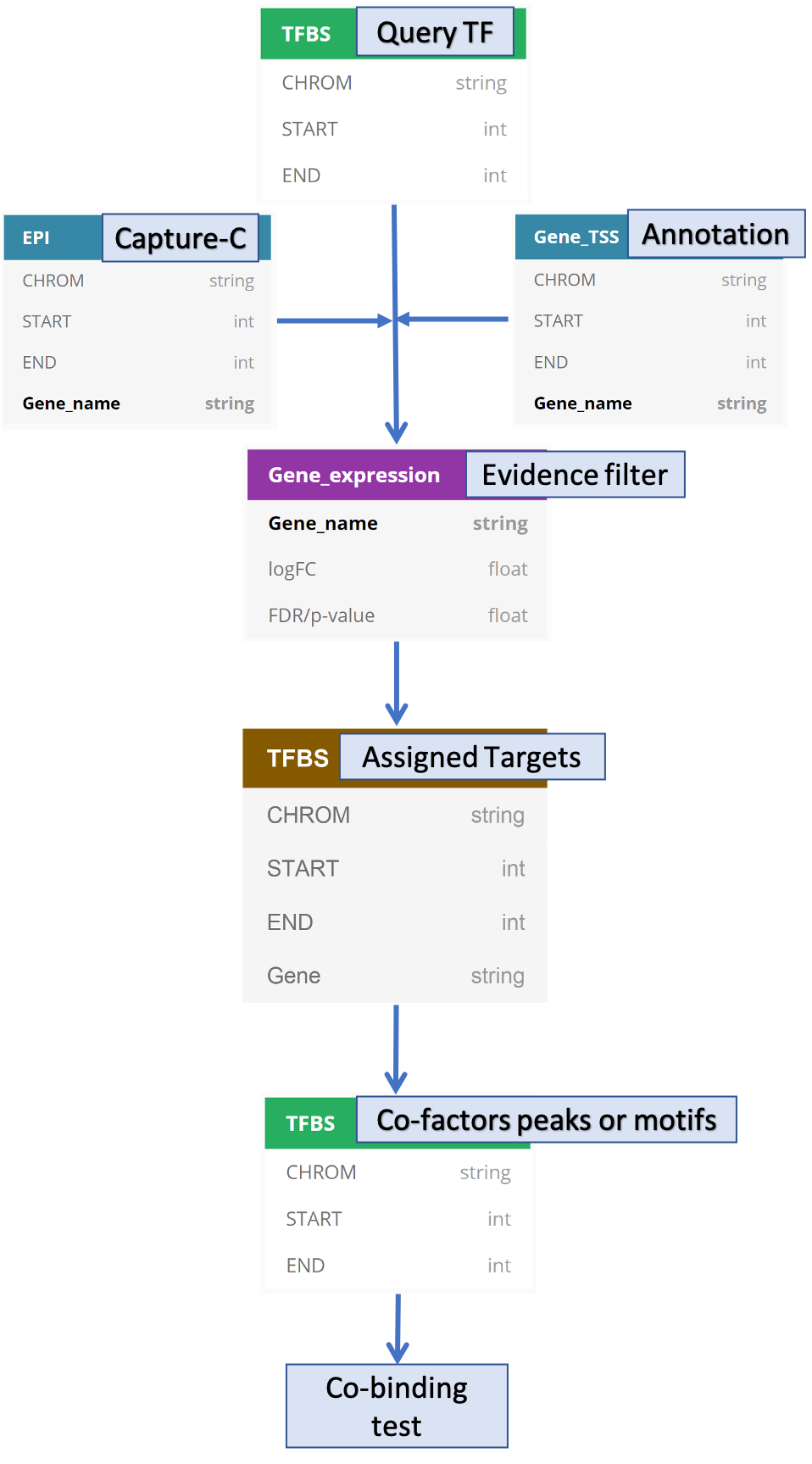
Input¶
query bed file (
required)
A tsv file. The first 3 columns should be chr, start, end. Additional columns will be ignored
tss annotation (default is mm9)
A tsv file. The first 4 columns should be chr, start, end, gene name. Additional columns will be ignored.
EPI data (default is mm9 HPC7 promoter capture HiC)
A tsv file. The first 4 columns should be chr, start, end, gene name. If 5th column is found, it will be used as interaction score. Additional columns will be ignored.
RNA-seq data (
required)
A tsv file with header, the first column should be gene name. User should specify LFC column name and FDR column name.
A list of chip-seq peaks used for co-binding test (default is mm9 HPC7 31 chip-seq datasets)
ERR1088371_Cebpb /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088371_Cebpb_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088372_cFos /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088372_cFos_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088373_cMyc /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088373_cMyc_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088378_E2f4 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088378_E2f4_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088379_Egr1 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088379_Egr1_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088380_Elf1 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088380_Elf1_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088381_Eto2 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088381_Eto2_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088382_Gata2 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088382_Gata2_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088383_H2A_AcK5 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088383_H2A_AcK5_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088384_H3K27me3 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088384_H3K27me3_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088385_H3K36me3 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088385_H3K36me3_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088386_H3K4me3 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088386_H3K4me3_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088409_Jun /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088409_Jun_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088410_Ldb1 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088410_Ldb1_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088411_Max /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088411_Max_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088412_Myb /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088412_Myb_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088413_Nfe2 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088413_Nfe2_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088414_p53 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088414_p53_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088415_Rad21 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088415_Rad21_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088416_Stat1P /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088416_Stat1P_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
ERR1088417_Stat3 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/ERR1088417_Stat3_HPC7.vs.ERR1088408_IgG_HPC7_peaks.rmblck.narrowPeak
SRR054909_GSM552232_H3AcK9 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/SRR054909_HPCminus7_Cell_Line_GSM552232_HPC7_H3AcK9_HPCminus7_Cell_Line.vs.SRR054913_HPCminus7_Cell_Line_GSM552236_HPC7_IgG_HPCminus7_Cell_Line_Input_peaks.rmblck.narrowPeak
SRR054910_GSM552233_Fli1 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/SRR054910_HPCminus7_Cell_Line_GSM552233_HPC7_Fli1_HPCminus7_Cell_Line.vs.SRR054913_HPCminus7_Cell_Line_GSM552236_HPC7_IgG_HPCminus7_Cell_Line_Input_peaks.rmblck.narrowPeak
SRR054911_GSM552234_Gata2 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/SRR054911_HPCminus7_Cell_Line_GSM552234_HPC7_Gata2_HPCminus7_Cell_Line.vs.SRR054913_HPCminus7_Cell_Line_GSM552236_HPC7_IgG_HPCminus7_Cell_Line_Input_peaks.rmblck.narrowPeak
SRR054912_GSM552235_Gfi1b /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/SRR054912_HPCminus7_Cell_Line_GSM552235_HPC7_Gfi1b_HPCminus7_Cell_Line.vs.SRR054913_HPCminus7_Cell_Line_GSM552236_HPC7_IgG_HPCminus7_Cell_Line_Input_peaks.rmblck.narrowPeak
SRR054914_GSM552237_Lmo2 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/SRR054914_HPCminus7_Cell_Line_GSM552237_HPC7_Lmo2_HPCminus7_Cell_Line.vs.SRR054913_HPCminus7_Cell_Line_GSM552236_HPC7_IgG_HPCminus7_Cell_Line_Input_peaks.rmblck.narrowPeak
SRR054915_GSM552238_Lyl1 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/SRR054915_HPCminus7_Cell_Line_GSM552238_HPC7_Lyl1_HPCminus7_Cell_Line.vs.SRR054913_HPCminus7_Cell_Line_GSM552236_HPC7_IgG_HPCminus7_Cell_Line_Input_peaks.rmblck.narrowPeak
SRR054916_GSM552239_Meis1 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/SRR054916_HPCminus7_Cell_Line_GSM552239_HPC7_Meis1_HPCminus7_Cell_Line.vs.SRR054913_HPCminus7_Cell_Line_GSM552236_HPC7_IgG_HPCminus7_Cell_Line_Input_peaks.rmblck.narrowPeak
SRR054917_GSM552240_Pu1 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/SRR054917_HPCminus7_Cell_Line_GSM552240_HPC7_Pu1_HPCminus7_Cell_Line.vs.SRR054913_HPCminus7_Cell_Line_GSM552236_HPC7_IgG_HPCminus7_Cell_Line_Input_peaks.rmblck.narrowPeak
SRR054918_GSM552241_Runx1 /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/SRR054918_HPCminus7_Cell_Line_GSM552241_HPC7_Runx1_HPCminus7_Cell_Line.vs.SRR054913_HPCminus7_Cell_Line_GSM552236_HPC7_IgG_HPCminus7_Cell_Line_Input_peaks.rmblck.narrowPeak
SRR054919_GSM552242_Scl /home/yli11/Tools/TF_target_finder/data/HPC7_chip_seq/SRR054919_HPCminus7_Cell_Line_GSM552242_HPC7_Scl_HPCminus7_Cell_Line.vs.SRR054913_HPCminus7_Cell_Line_GSM552236_HPC7_IgG_HPCminus7_Cell_Line_Input_peaks.rmblck.narrowPeak
6.a. main TF motif pwm files (required)
/home/yli11/Tools/TF_target_finder/data/NFIX_mouse_known_motifs.meme
6.b. A list of motif ids used for co-binding test (default is selected mouse motifs)
This input is a tsv file containing TF name and motif names (separated by comma). Full mapping file can be found at: motif mapping table
CEBPB CEBPB_MOUSE.H11MO.0.A,M0314_1.02
CMYC CMYC
E2F4 E2F4_MOUSE.H11MO.0.A,E2F4_MOUSE.H11MO.1.A,M4537_1.02
EGR1 EGR1_MOUSE.H11MO.0.A,M0417_1.02,UP00007_1,UP00007_2
ELF1 ELF1_MOUSE.H11MO.0.A,M4688_1.02
FLI1 FLI1_MOUSE.H11MO.0.A,FLI1_MOUSE.H11MO.1.A,M0699_1.02
GATA2 GATA2_MOUSE.H11MO.0.A,M4660_1.02
JUN JUN_MOUSE.H11MO.0.A,JUNB_MOUSE.H11MO.0.A,JUND_MOUSE.H11MO.0.A,M0311_1.02,M0312_1.02,M0320_1.02,UP00103_1,UP00103_2
LYL1 LYL1_MOUSE.H11MO.0.A
MAX M0221_1.02,MAX_MOUSE.H11MO.0.A,UP00060_1,UP00060_2
MEIS1 M2298_1.02,MEIS1_MOUSE.H11MO.0.A,MEIS1_MOUSE.H11MO.1.A,UP00186_1
MYB M1923_1.02,MYB_MOUSE.H11MO.0.A,MYBA_MOUSE.H11MO.0.C,MYBB_MOUSE.H11MO.0.D
NFE2 M4629_1.02,M6359_1.02,NFE2_MOUSE.H11MO.0.A
P53 P53_MOUSE.H11MO.0.A,P53_MOUSE.H11MO.1.A
RUNX1 M1837_1.02,RUNX1_MOUSE.H11MO.0.A
PU.1 SPI1_MOUSE.H11MO.0.A,UP00085_1,UP00085_2,M6122_1.02
STAT3 STAT3,STAT3_MOUSE.H11MO.0.A
STAT1 STAT1_MOUSE.H11MO.0.A,STAT1_MOUSE.H11MO.1.A
TAL1 TAL1_MOUSE.H11MO.0.A
GFI1B GFI1B_MOUSE.H11MO.0.A
Usage¶
hpcf_interative
module load python/2.7.13
tf_target_finder.py --label NFIX -q NFIX_idr_peaks.bed -exp results.KO_vs_WT.txt --query_motif /home/yli11/Tools/TF_target_finder/data/NFIX_mouse_known_motifs.meme
Output¶
Inside the jobID folder, you can find:
assign_targets.log: statistics of target assignments
[label].query.DEG_targets_filter.bed: subset of query file with targets assigned
[label].query.targets_all.bed: query file with candidate targets as additional column
[label].deg_table.tsv: subset of deg table on candidate targets
assign_targets_output.tsv: query file with additional columns, including nearest TSS, gene within TSS flank, EPI assigned targets and associated scores
Results of motif co-binding test:
motif_co_binding_test/motif_summary.txtResults of peak co-binding test:
peak_co_binding_test/motif_summary.txt
Comments¶
code @ github.