Single-end ChIP-seq¶
usage: HemTools chip_seq_single [-h] [-j JID] [--short] [--debug] [-f INPUT]
[-d DESIGN_MATRIX] [--guess_input]
[-i INDEX_FILE] [-g GENOME] [-b BLACKLIST]
[-s CHROM_SIZE] [-e EFFECTIVEGENOMESIZE]
Named Arguments¶
- -j, --jid
enter a job ID, which is used to make a new directory. Every output will be moved into this folder.
Default: “{{subcmd}}_docs_2024-04-18”
- --short
Force to use the short queue. (only if R1+R2 fastq.gz size <=250M)
Default: False
- --debug
Not for end-user.
Default: False
- -f, --input
tab delimited 3 columns (tsv file): Read 1 fastq, Read 2 fastq, sample ID
- -d, --design_matrix
tab delimited 3 columns (tsv file): treatment sample ID, control sample ID, peakcall ID
- --guess_input
Let the program generate the input files for you.
Default: False
Genome Info¶
- -i, --index_file
BWA index file
Default: “/home/docs/checkouts/readthedocs.org/user_builds/hemtools/checkouts/latest/subcmd/../hg19/bwa_16a_index/hg19.fa”
- -g, --genome
genome version: hg19, hg38, mm10, mm9.
Default: “hg19”
- -b, --Blacklist
Blacklist file
Default: “/home/docs/checkouts/readthedocs.org/user_builds/hemtools/checkouts/latest/subcmd/../hg19/Hg19_Blacklist.bed”
- -s, --chrom_size
chrome size
Default: “/home/docs/checkouts/readthedocs.org/user_builds/hemtools/checkouts/latest/subcmd/../hg19/hg19.chrom.sizes”
- -e, --effectiveGenomeSize
effectiveGenomeSize for bamCoverage
Default: “2451960000”
Usage¶
Go to your data directory and type the following.
Step 0: Load python version 2.7.13.
$ module load python/2.7.13
Step 1: Prepare input files, generate fastq.tsv.
$ HemTools chip_seq_single --guess_input
Input fastq files preparation complete! ALL GOOD!
Please check if you like the computer-generated labels in : fastq.tsv
Input peakcall file preparation complete! File name: peakcall.tsv
Note
If you are preparing fastq.tsv and peakcall.tsv yourself, please make sure no space anywhere in the file. Note that the seperator is tab. Spaces in file name will cause errors.
Step 2: Check the computer-generated input list (manually), make sure they are correct.
$ less fastq.tsv
$ less peakcall.tsv
Note
a random string will be added to the generated files (e.g., fastq.94c049cbff1f.tsv) if they exist before running step 1.
Step 3: Submit your job.
$ HemTools chip_seq_single -f fastq.tsv -d peakcall.tsv
Sample input format¶
fastq.tsv
This is a tab-seperated-value format file. The 2 columns are: Read 1, sample ID.
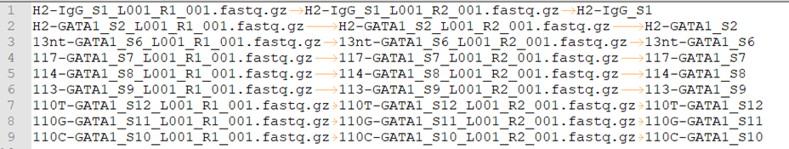
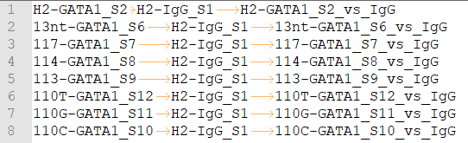
peakcall.tsv
This is also a tab-seperated-value format file. The 3 columns are: treatment sample ID, control/input sample ID, peakcall ID.
Quality Control¶
The quality metrics are provided in the html report. For ChIP-seq data, we also provide strand cross-correlation metrics (i.e., those attached pdf files).
Metrics |
Threshold |
NRF |
>0.9 |
PBC1 |
>0.9 |
PBC2 |
>10 |
RSC |
>0.8 |
QTag |
>=1 |
https://www.encodeproject.org/atac-seq/
https://www.encodeproject.org/chip-seq/transcription_factor/ https://www.encodeproject.org/chip-seq/histone/
https://github.com/crazyhottommy/ChIP-seq-analysis/blob/master/part0_quality_control.md
Report bug¶
Once the job is finished, you will be notified by email with some attachments. If no attachment can be found, it might be caused by an error. In such case, please go to the result directory (where the log_files folder is located) and type:
$ HemTools report_bug
Use different genome index¶
$ HemTools chip_seq_single -f fastq.tsv -d peakcall.tsv -i YOUR_GENOME_INDEX
Comments¶
code @ github.